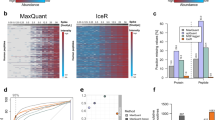
Abstract
Consistent detection and quantification of protein post-translational modifications (PTMs) across sample cohorts is a prerequisite for functional analysis of biological processes. Data-independent acquisition (DIA) is a bottom-up mass spectrometry approach that provides complete information on precursor and fragment ions. However, owing to the convoluted structure of DIA data sets, confident, systematic identification and quantification of peptidoforms has remained challenging. Here, we present inference of peptidoforms (IPF), a fully automated algorithm that uses spectral libraries to query, validate and quantify peptidoforms in DIA data sets. The method was developed on data acquired by the DIA method SWATH-MS and benchmarked using a synthetic phosphopeptide reference data set and phosphopeptide-enriched samples. IPF reduced false site-localization by more than sevenfold compared with previous approaches, while recovering 85.4% of the true signals. Using IPF, we quantified peptidoforms in DIA data acquired from >200 samples of blood plasma of a human twin cohort and assessed the contribution of heritable, environmental and longitudinal effects on their PTMs.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Deutsch, E.W. et al. State of the human proteome in 2014/2015 as viewed through PeptideAtlas: enhancing accuracy and coverage through the AtlasProphet. J. Proteome Res. 14, 3461–3473 (2015).
Smith, L.M. & Kelleher, N.L. Proteoform: a single term describing protein complexity. Nat. Methods 10, 186–187 (2013).
Lane, L. et al. neXtProt: a knowledge platform for human proteins. Nucleic Acids Res. 40, D76–D83 (2012).
Uhlén, M. et al. Proteomics. Tissue-based map of the human proteome. Science 347, 1260419 (2015).
Edwards, A.M. et al. Too many roads not taken. Nature 470, 163–165 (2011).
Marx, V. Finding the right antibody for the job. Nat. Methods 10, 703–707 (2013).
Chait, B.T. Chemistry. Mass spectrometry: bottom-up or top-down? Science 314, 65–66 (2006).
Aebersold, R. & Mann, M. Mass spectrometry-based proteomics. Nature 422, 198–207 (2003).
Aebersold, R. & Mann, M. Mass-spectrometric exploration of proteome structure and function. Nature 537, 347–355 (2016).
Domon, B. & Aebersold, R. Options and considerations when selecting a quantitative proteomics strategy. Nat. Biotechnol. 28, 710–721 (2010).
Picotti, P. & Aebersold, R. Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nat. Methods 9, 555–566 (2012).
Peterson, A.C., Russell, J.D., Bailey, D.J., Westphall, M.S. & Coon, J.J. Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Mol. Cell. Proteomics 11, 1475–1488 (2012).
Chapman, J.D., Goodlett, D.R. & Masselon, C.D. Multiplexed and data-independent tandem mass spectrometry for global proteome profiling. Mass Spectrom. Rev. 33, 452–470 (2014).
Gillet, L.C. et al. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol. Cell. Proteomics 11, 016717 (2012).
Navarro, P. et al. A multicenter study benchmarks software tools for label-free proteome quantification. Nat. Biotechnol. 34, 1130–1136 (2016).
Doerr, A. DIA mass spectrometry. Nat. Methods 12, 35 (2015).
Na, S. & Paek, E. Software eyes for protein post-translational modifications. Mass Spectrom. Rev. 34, 133–147 (2015).
Chalkley, R.J. & Clauser, K.R. Modification site localization scoring: strategies and performance. Mol. Cell. Proteomics 11, 3–14 (2012).
Oliveira, A.P. et al. Regulation of yeast central metabolism by enzyme phosphorylation. Mol. Syst. Biol. 8, 623 (2012).
Abelin, J.G. et al. Reduced-representation phosphosignatures measured by quantitative targeted ms capture cellular states and enable large-scale comparison of drug-induced phenotypes. Mol. Cell. Proteomics 15, 1622–1641 (2016).
Silva, J.C. et al. Quantitative proteomic analysis by accurate mass retention time pairs. Anal. Chem. 77, 2187–2200 (2005).
Tsou, C.C. et al. DIA-Umpire: comprehensive computational framework for data-independent acquisition proteomics. Nat. Methods 12, 258–264, 7, 264 (2015).
Li, Y. et al. Group-DIA: analyzing multiple data-independent acquisition mass spectrometry data files. Nat. Methods 12, 1105–1106 (2015).
Wang, J. et al. MSPLIT-DIA: sensitive peptide identification for data-independent acquisition. Nat. Methods 12, 1106–1108 (2015).
Ting, Y.S. et al. Peptide-centric proteome analysis: an alternative strategy for the analysis of tandem mass spectrometry data. Mol. Cell. Proteomics 14, 2301–2307 (2015).
Röst, H.L. et al. OpenSWATH enables automated, targeted analysis of data-independent acquisition MS data. Nat. Biotechnol. 32, 219–223 (2014).
Sidoli, S. et al. Sequential window acquisition of all theoretical mass spectra (SWATH) analysis for characterization and quantification of histone post-translational modifications. Mol. Cell. Proteomics 14, 2420–2428 (2015).
Krautkramer, K.A., Reiter, L., Denu, J.M. & Dowell, J.A. Quantification of SAHA-dependent changes in histone modifications using data-independent acquisition mass spectrometry. J. Proteome Res. 14, 3252–3262 (2015).
Porter, C.J. & Bereman, M.S. Data-independent-acquisition mass spectrometry for identification of targeted-peptide site-specific modifications. Anal. Bioanal. Chem. 407, 6627–6635 (2015).
Lawrence, R.T., Searle, B.C., Llovet, A. & Villén, J. Plug-and-play analysis of the human phosphoproteome by targeted high-resolution mass spectrometry. Nat. Methods 13, 431–434 (2016).
Keller, A. et al. Opening a SWATH window on posttranslational modifications: automated pursuit of modified peptides. Mol. Cell. Proteomics 15, 1151–1163 (2016).
Schubert, O.T. et al. Building high-quality assay libraries for targeted analysis of SWATH MS data. Nat. Protoc. 10, 426–441 (2015).
Carr, S.A. et al. Targeted peptide measurements in biology and medicine: best practices for mass spectrometry-based assay development using a fit-for-purpose approach. Mol. Cell. Proteomics 13, 907–917 (2014).
Röst, H.L. et al. TRIC: an automated alignment strategy for reproducible protein quantification in targeted proteomics. Nat. Methods 13, 777–783 (2016).
Soste, M. et al. A sentinel protein assay for simultaneously quantifying cellular processes. Nat. Methods 11, 1045–1048 (2014).
Choi, H., Ghosh, D. & Nesvizhskii, A.I. Statistical validation of peptide identifications in large-scale proteomics using the target-decoy database search strategy and flexible mixture modeling. J. Proteome Res. 7, 286–292 (2008).
Choi, H. & Nesvizhskii, A.I. Semisupervised model-based validation of peptide identifications in mass spectrometry-based proteomics. J. Proteome Res. 7, 254–265 (2008).
Fermin, D., Walmsley, S.J., Gingras, A.-C., Choi, H. & Nesvizhskii, A.I. LuciPHOr: algorithm for phosphorylation site localization with false localization rate estimation using modified target-decoy approach. Mol. Cell. Proteomics 12, 3409–3419 (2013).
Fermin, D., Avtonomov, D., Choi, H. & Nesvizhskii, A.I. LuciPHOr2: site localization of generic post-translational modifications from tandem mass spectrometry data. Bioinformatics 31, 1141–1143 (2015).
Nagano, K. et al. Phosphoproteomic analysis of distinct tumor cell lines in response to nocodazole treatment. Proteomics 9, 2861–2874 (2009).
Teo, G. et al. mapDIA: Preprocessing and statistical analysis of quantitative proteomics data from data independent acquisition mass spectrometry. J. Proteomics 129, 108–120 (2015).
Collins, B.C. et al. Quantifying protein interaction dynamics by SWATH mass spectrometry: application to the 14-3-3 system. Nat. Methods 10, 1246–1253 (2013).
Liu, Y. et al. Quantitative variability of 342 plasma proteins in a human twin population. Mol. Syst. Biol. 11, 786 (2015).
Zhu, P., Bowden, P., Zhang, D. & Marshall, J.G. Mass spectrometry of peptides and proteins from human blood. Mass Spectrom. Rev. 30, 685–732 (2011).
Yang, H. & Zubarev, R.A. Mass spectrometric analysis of asparagine deamidation and aspartate isomerization in polypeptides. Electrophoresis 31, 1764–1772 (2010).
Zawadzka, A.M. et al. Variation and quantification among a target set of phosphopeptides in human plasma by multiple reaction monitoring and SWATH-MS2 data-independent acquisition. Electrophoresis 35, 3487–3497 (2014).
Abello, N., Kerstjens, H.A.M., Postma, D.S. & Bischoff, R. Protein tyrosine nitration: selectivity, physicochemical and biological consequences, denitration, and proteomics methods for the identification of tyrosine-nitrated proteins. J. Proteome Res. 8, 3222–3238 (2009).
Gu, H. et al. Quantitative profiling of post-translational modifications by immunoaffinity enrichment and LC-MS/MS in cancer serum without immunodepletion. Mol. Cell. Proteomics 15, 692–702 (2016).
Hüttenhain, R. et al. Reproducible quantification of cancer-associated proteins in body fluids using targeted proteomics. Sci. Transl. Med. 4, 142ra94 (2012).
Anderson, N.L. & Anderson, N.G. The human plasma proteome: history, character, and diagnostic prospects. Mol. Cell. Proteomics 1, 845–867 (2002).
Huang, Y. et al. Myeloperoxidase, paraoxonase-1, and HDL form a functional ternary complex. J. Clin. Invest. 123, 3815–3828 (2013).
Huang, Y. et al. An abundant dysfunctional apolipoprotein A1 in human atheroma. Nat. Med. 20, 193–203 (2014).
Borhani, D.W., Rogers, D.P., Engler, J.A. & Brouillette, C.G. Crystal structure of truncated human apolipoprotein A-I suggests a lipid-bound conformation. Proc. Natl. Acad. Sci. USA 94, 12291–12296 (1997).
Pankhurst, G. et al. Characterization of specifically oxidized apolipoproteins in mildly oxidized high density lipoprotein. J. Lipid Res. 44, 349–355 (2003).
Egertson, J.D. et al. Multiplexed MS/MS for improved data-independent acquisition. Nat. Methods 10, 744–746 (2013).
Tsou, C.-C., Tsai, C.F., Teo, G.C., Chen, Y.J. & Nesvizhskii, A.I. Untargeted, spectral library-free analysis of data-independent acquisition proteomics data generated using Orbitrap mass spectrometers. Proteomics 16, 2257–2271 (2016).
Liu, T. et al. Evaluation of multiprotein immunoaffinity subtraction for plasma proteomics and candidate biomarker discovery using mass spectrometry. Mol. Cell. Proteomics 5, 2167–2174 (2006).
Kim, S.C. et al. A clean, more efficient method for in-solution digestion of protein mixtures without detergent or urea. J. Proteome Res. 5, 3446–3452 (2006).
Escher, C. et al. Using iRT, a normalized retention time for more targeted measurement of peptides. Proteomics 12, 1111–1121 (2012).
Liu, Y. et al. Quantitative measurements of N-linked glycoproteins in human plasma by SWATH-MS. Proteomics 13, 1247–1256 (2013).
Chambers, M.C. et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 30, 918–920 (2012).
Eng, J.K., Jahan, T.A. & Hoopmann, M.R. Comet: an open-source MS/MS sequence database search tool. Proteomics 13, 22–24 (2013).
Keller, A., Nesvizhskii, A.I., Kolker, E. & Aebersold, R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 (2002).
Deutsch, E.W. et al. A guided tour of the Trans-Proteomic Pipeline. Proteomics 10, 1150–1159 (2010).
Shteynberg, D. et al. iProphet: multi-level integrative analysis of shotgun proteomic data improves peptide and protein identification rates and error estimates. Mol. Cell. Proteomics 10, 007690 (2011).
Lam, H. et al. Building consensus spectral libraries for peptide identification in proteomics. Nat. Methods 5, 873–875 (2008).
Röst, H.L. et al. OpenMS: a flexible open-source software platform for mass spectrometry data analysis. Nat. Methods 13, 741–748 (2016).
Cherry, J.M. et al. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 40, D700–D705 (2012).
Magrane, M. & UniProt Consortium UniProt Knowledgebase: a hub of integrated protein data. Database (Oxford) 2011, bar009 (2011).
Zhou, H. et al. Robust phosphoproteome enrichment using monodisperse microsphere-based immobilized titanium (IV) ion affinity chromatography. Nat. Protoc. 8, 461–480 (2013).
Tyanova, S., Temu, T. & Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11, 2301–2319 (2016).
Almasy, L. & Blangero, J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am. J. Hum. Genet. 62, 1198–1211 (1998).
Storey, J.D. & Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100, 9440–9445 (2003).
Teleman, J. et al. DIANA--algorithmic improvements for analysis of data-independent acquisition MS data. Bioinformatics 31, 555–562 (2015).
Vizcaíno, J.A. et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44 D1, D447–D456 (2016).
Acknowledgements
H.L.R. was funded by the Swiss National Science Foundation (SNSF grant P2EZP3 162268). R.A. was supported by ERC Proteomics v3.0 (AdG-233226 Proteomics v.3.0 and AdG-670821 Proteomics 4D) and the Swiss National Science Foundation (SNSF) (31003A_166435). We would like to thank L. Gillet and A. Leitner for insightful discussions on post-translational modification and SWATH-MS. We are grateful to all twin registry participants recruited in this study. For the unit of Twins UK, this study was funded by the Wellcome Trust and EC's Seventh Framework Programme (FP7/2007-2013) and also received support from the National Institute for Health Research-funded BioResource, Clinical Research Facility and Biomedical Research Centre based at Guy's and St Thomas' NHS Foundation Trust in partnership with King's College London. Further acknowledgments go to the Scientific IT Support team of ETH Zurich for support and maintenance of the lab-internal computing infrastructure, the HPC team (Brutus) and the OpenMS and PyProphet developers for including IPF in the OpenMS and PyProphet frameworks. We thank the PRIDE team for proteomic data deposition.
Author information
Authors and Affiliations
Contributions
G.R. developed and implemented IPF and analyzed the synthetic phosphopeptide reference, enriched phosphopeptide, 14-3-3β and twin study data. Y.L. provided and analyzed the twin study data. H.L.R. developed and implemented the MS1 scoring and quantification in OpenSWATH. C.L. provided the synthetic phosphopeptide reference sample and acquired the data. A. Buil and G.R. conducted the heritability analysis of the twin study. A. Bensimon and Y.L. conducted the enriched phosphopeptide experiment and acquired the data. M.S. provided the synthetic phosphopeptide reference sample. B.C.C. analyzed the 14-3-3β data. T.D.S. and E.T.D. designed and supervised the twin study. All authors provided critical input on the project. G.R., Y.L. and R.A. wrote the paper with feedback from all authors. L.M. supervised the development of IPF and conducted the protein-level PTM meta-analysis. R.A. designed and supervised the study.
Corresponding author
Ethics declarations
Competing interests
R.A. holds shares of Biognosys AG, which operates in the field covered by the article. The remaining authors declare no competing financial interest.
Integrated supplementary information
Supplementary Figure 1 IPF analysis workflow overview.
a) Query parameter generation: Based on a discovery proteomics workflow (DDA or DIA), peptide query parameters consisting of detection and identification transitions for all peptidoforms are generated. b) Signal processing: Using a two-tier scoring approach, the detection and identification transitions are extracted from the SWATH maps. The detection transitions are used to find candidate peak groups against which the identification transitions are scored. The two-tier scoring estimates posterior probabilities (PPs) for the candidate peak groups using the detection transitions. In the step on transition-level, the PPs that individual identification transitions are originating from the peak group associated peptide are estimated. c) Statistical inference: A Bayesian hierarchical model (BHM) integrates the transition PPs according to residue specificity to peptidoform PPs. In addition to the peptidoforms, the PP that the signal is a false positive (h0) is being updated. In data sets with large precursor isolation windows, the BHM is extended by an intermediate layer to adjust the probabilities using precursor data on MS1 and MS2-level data (see Methods).
Supplementary Figure 2 Discriminant score histograms on MS1-, MS2- and transition-levels used for multi-tier scoring on the synthetic phosphopeptide reference data set.
Instead of assuming a normal distribution for false targets and decoys, a non-parametric model is used on all levels. a) On MS1-level, only a small fraction of the targets can be detected with confidence. b) MS2-level feature detection enables more sensitive peak group detection than the MS2-guided MS1 scoring. c) Only a small fraction of the target transitions can be detected by the transition scoring after MS2 scoring (peptide query FDR 1%) because the number of theoretical transitions exceeds by far the number of detectable fragment ions.
Supplementary Figure 3 Benchmarking on the synthetic phosphopeptide reference data set using the DIA-Umpire library.
Spiked-in synthetic yeast phosphopeptides were measured in a 13-step dilution series with a human cell line background. IPF was applied using peptide query parameters generated from a combined spectral library based on the DIA-Umpire analysis of the 13 runs. a) The receiver operating characteristic (ROC) indicates high sensitivity at commonly used confidence thresholds with 73.1% recovery at 5% (grey dotted line) false positive rate. b) The estimated global false discovery rate (FDR) or q-values are plotted against the true FDR, computed using the ground truth. The dashed diagonal line indicates the optimum. The estimated local false discovery rate (fdr) or posterior error probability (PEP) is plotted against the actual fdr, approximated using the ground truth as described previously with a window size of 500. IPF enables accurate estimation in the commonly used ranges of 1-5% fdr/FDR, with a small overestimation of the error in the higher ranges. c) The dilution series of synthetic spiked-in standard peptides against the constant human cell line and the detected true (green) and false (red) peptidoforms at 5% FDR are depicted. The light colors (OSW) represent the detectable peptide sequence-specific peptide query-level MS2 signals. The dark colors (IPF) represent the corrected, peptidoform-specific signals. A high gain in selectivity with a small drop in sensitivity can be observed. d) The quantification of the peak groups (normalized against 1:0) is compared against the ground truth (red line). Until dilution step 1:15 the quantification is accurate, with a slight bias for overestimation at lower abundance dilution steps. The numbers above the boxplots indicate the number of peptides per dilution steps that are also present in the 1:0 step. e) The boxplots depict the intensities of correct peptidoforms and background (BG) peptides at 5% FDR. To achieve high confidence on peptidoform-level, IPF requires slightly higher signal intensities than OpenSWATH on peptide-level.
Supplementary Figure 4 Comparison of false localization rates estimated by IPF and DIA-Umpire/LuciPHOr.
The synthetic phosphopeptide reference data set analysed with DIA-Umpire was benchmarked using IPF and DIA-Umpire/LuciPHOr. The IPF analysis was conducted using the DIA-Umpire spectral library and using a simplified Bayesian hierarchical model which estimated a false localization rate comparable to the metric reported by LuciPHOr. a) The pseudo receiver operating characteristics (ROC) for both IPF (red) and DIA-Umpire/LuciPHOr (cyan) indicate high sensitivity and specificity at 5% (grey dotted line) false positive rate (FPR) with IPF achieving a higher true positive rate (TPR). Because neither IPF nor DIA-Umpire/LuciPHOr can identify all peptide site-localizations in all samples, they do not reach a true positive rate (TPR) of 1.0. While the TPR of LuciPHOr saturates at above 0.6, for IPF the TPR goes above 0.8, indicating higher sensitivity at the same FPR. b) Both IPF (red) and DIA-Umpire/LuciPHOr (cyan) accurately estimate global (FLR) and local (flr) false localization rates. However, the FLR/flr estimates of DIA-Umpire/LuciPHOr are impaired by false positive identifications at high confidence thresholds below 1% FLR/flr. c-d) The dilution series of synthetic spiked-in standard peptides against the constant human cell line and the detected true (solid) and false (dashed) peptide site-localizations at 5% and 10% FLR are depicted. In general, IPF (red) enables higher numbers of correct site-localizations across the dilution series than DIA-Umpire/LuciPHOr (cyan), with the effect being more pronounced at 10% FLR.
Supplementary Figure 5 Benchmarking using phosphopeptide-enriched samples.
Enriched phosphopeptide samples of a human U2OS cell line treated with nocodazole and without treatment (control) were measured in both DDA and DIA modes in each 10 replicates. a) 200 peptides were randomly selected and the corresponding detected peak groups and peptide precursors for IPF and MaxQuant (MQ) are visualized in a heatmap (sorted by a hierarchical dendrogram for identification/detection by rows) for detectability/identification confidence (blue) and quantification (red-yellow; including alignment). IPF achieved a higher level of completeness for quantification in individual experimental conditions (Nocodazole N01-N10: 62.6%; Controls C01-C10: 47.5%) in comparison to MaxQuant (Nocodazole N01-N10: 22.9%; Controls C01-C10: 16.5%). b) The consistency of quantification for all intersecting peptides is depicted, where IPF provided more complete detection in replicates than MaxQuant. c) Differential expression analysis was conducted using mapDIA (significance thresholds: FDR < 0.01 & log2(FC) > 2). For both MaxQuant and IPF on MS2 peak group (IPF-MS2) and MS1 (IPF-MS1) precursor levels, the same peptide/precursor-level model and parameters were used.
Supplementary Figure 6 Quantification of phosphorylation dynamics in the 14-3-3 system.
a) Across the full time series, IPF detected and quantified a higher number of phosphopeptides (43.6%) in two or more replicates than MaxQuant (9.5%). b-c) Phosphopeptides associated with the 14-3-3 binding motif are expected to correlate in abundance with their associated prey proteins. The correlation of the quantitative profiles across the full time series of consistently measured phosphopeptides (in or outside the motif) was computed and is depicted as boxplots. The number within the boxes indicate the number of data points. IPF achieves a higher absolute correlation than MaxQuant.
Supplementary Figure 7 Peptidoform dynamic range of top 100 most abundant proteins.
The smoothed scatter plot of peptidoform quantile normalized intensity (detected with q-value < 0.01) over all samples is depicted.
Supplementary Figure 8 Peptidoform dynamic range of lower abundant proteins.
The smoothed scatter plot of peptidoform quantile normalized intensity (detected with q-value < 0.01) over all samples is depicted.
Supplementary Figure 9 Detectability and quantification of modified peptides in technical replicates of human blood plasma.
The detectability in number of technical replicates across different modification types and the coefficient of variation (CV; computed only if the peptidoform was detected in at least 2 runs) of the quantile normalized peak group intensities across different modification types is depicted. Most modified peptidoforms could consistently be detected in the majority of the technical replicates. The coefficient of variation of the intensities is commonly within the expected range (10-20%) of SWATH-MS.
Supplementary Figure 10 Detectability and quantification of modified peptides in whole-process replicates of human blood plasma.
The detectability in number of whole-process replicates across different modification types and the coefficient of variation (CV; computed only if the peptidoform was detected in at least 2 runs) of the quantile normalized peak group intensities across different modification types is depicted. Most modified peptidoforms could consistently be detected in the majority of the whole-process replicates. The coefficient of variation of the peak group intensity is higher than for the technical replicates, representing the additional introduced variability of the sample preparation and processing steps.
Supplementary Figure 11 ALBU modification type and peptidoform intensities distribution.
The quantitative variability (mean±sd) of peptidoform quantile normalized intensity (detected with q-value < 0.01) over all samples is depicted. While the peptidoform intensity cannot be used for absolute label-free quantification, peptidoforms carrying modifications are in general distributed over the whole intensity range, which might indicate that post-translationally modified ALBU proteoforms are substantial components of the total ALBU protein mass.
Supplementary Figure 12 Longitudinal ALBU abundance fold change.
The boxplot depicts the peptidoform abundance fold changes of all ALBU peptidoforms between time points 2 (later) and 1 (log2(NI_2/NI_1); NI: quantile normalized intensity) for all individuals. The relative ALBU peptidoform abundances remain similar over time with only a slow decrease.
Supplementary Figure 13 Dissection of the plasma peptidoform-level variability.
The relative variance components (RVC) are plotted in a scatterplot against the -log10(q-value) per peptidoform peak group. The black dashed lines indicate significance thresholds (q-value < 0.01). The RVC of an individual peptidoform is affected by several different factors, most importantly the protein abundance and proteoform variability as well as technical effects. a-b) Peptides of ApoE allele variants E2 (green circles) and E3/E4 (red circles) (wild-type) were detected and quantified in the samples (Supplementary Note V.D). The heritable and common environment effects are often accounted together as “family” effects because discrimination of the two is difficult. For the E2 allele, the heritable component was found to amount for 56.2% of the variance (c2: 13.5% h2id: 8.1%, h2w: 3.7%, e2: 18.5%). For the E3/E4 allele, the heritable component was found to amount for 17.6% of the variance (c2: 34.9% h2id: 19.2%, h2w: 1.5%, e2: 26.8%). c) The longitudinal component was found to be the major contributor to ApoA1 tryptophan oxidation (orange circles) abundance variance.
Supplementary Figure 14 oxMet86 peptidoform abundance fold changes.
The boxplots depict the peptidoform abundance fold changes between time points 2 (later) and 1 (log2(NI_2/NI_1); NI: quantile normalized intensity) for all individuals. The methionine oxidized peptidoforms (oxMet86) show a decrease, which might be induced by the longer time of sample storage and thus spontaneous methionine oxidation for the samples at the first visits.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–14 and Supplementary Notes (PDF 9627 kb)
Supplementary Table 1
Synthetic phosphopeptide reference data set: Peptide sequences (TXT 24 kb)
Supplementary Table 2
Twin plasma data set: Summary statistics of spectral library coverage and IPF detectability (CSV 0 kb)
Supplementary Table 3
Twin plasma data set: Quantitative variance components of peptidoforms (CSV 2856 kb)
Supplementary Table 4
Twin plasma data set: Peptide coverage of proteins (CSV 20 kb)
Supplementary Data 1
Twin plasma data set: Protein-level graphical reports (ZIP 1202 kb)
Rights and permissions
About this article
Cite this article
Rosenberger, G., Liu, Y., Röst, H. et al. Inference and quantification of peptidoforms in large sample cohorts by SWATH-MS. Nat Biotechnol 35, 781–788 (2017). https://doi.org/10.1038/nbt.3908
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.3908
This article is cited by
-
DeepFLR facilitates false localization rate control in phosphoproteomics
Nature Communications (2023)
-
An optogenetic-phosphoproteomic study reveals dynamic Akt1 signaling profiles in endothelial cells
Nature Communications (2023)
-
A data-independent acquisition-based global phosphoproteomics system enables deep profiling
Nature Communications (2021)
-
DeepPhospho accelerates DIA phosphoproteome profiling through in silico library generation
Nature Communications (2021)
-
GproDIA enables data-independent acquisition glycoproteomics with comprehensive statistical control
Nature Communications (2021)