Abstract
Targeted base editing in plants without the need for a foreign DNA donor or double-stranded DNA cleavage would accelerate genome modification and breeding in a wide array of crops. We used a CRISPR–Cas9 nickase-cytidine deaminase fusion to achieve targeted conversion of cytosine to thymine from position 3 to 9 within the protospacer in both protoplasts and regenerated rice, wheat and maize plants at frequencies of up to 43.48%.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Accession codes
Primary accessions
NCBI Reference Sequence
Sequence Read Archive
References
Voytas, D.F. & Gao, C. PLoS Biol. 12, e1001877 (2014).
Zhao, K. et al. Nat. Commun. 2, 467 (2011).
Henikoff, S. & Comai, L. Annu. Rev. Plant Biol. 54, 375–401 (2003).
Henikoff, S., Till, B.J. & Comai, L. Plant Physiol. 135, 630–636 (2004).
Slade, A.J., Fuerstenberg, S.I., Loeffler, D., Steine, M.N. & Facciotti, D. Nat. Biotechnol. 23, 75–81 (2005).
Hsu, P.D., Lander, E.S. & Zhang, F. Cell 157, 1262–1278 (2014).
Svitashev, S. et al. Plant Physiol. 169, 931–945 (2015).
Čermák, T., Baltes, N.J., Čegan, R., Zhang, Y. & Voytas, D.F. Genome Biol. 16, 232 (2015).
Komor, A.C., Kim, Y.B., Packer, M.S., Zuris, J.A. & Liu, D.R. Nature 533, 420–424 (2016).
Nishida, K. et al. Science http://dx.doi.org/10.1126/science.aaf8729 (2016).
Ma, Y. et al. Nat. Methods 13, 1029–1035 (2016).
Hess, G.T. et al. Nat. Methods 13, 1036–1042 (2016).
Feng, B. et al. J. Cereal Sci. 52, 387–394 (2010).
Zhong, C.X. et al. Plant Cell 14, 2825–2836 (2002).
Huang, Q.N. et al. J. Integr. Plant Biol. 58, 12–28 (2016).
Zhang, Y. et al. Nat. Commun. 7, 12617 (2016).
Lu, Y. & Zhu, J.K. Mol. Plant http://dx.doi.org/10.1016/j.molp.2016.11.013 (2016).
Li, J., Sun, Y., Du, J., Zhao, Y. & Xia, L. et al. Mol. Plant http://dx.doi.org/10.1016/j.molp.2016.12.001 (2016).
Ren, B. et al. Sci. China Life Sci. http://engine.scichina.com/doi/10.1007/s11427-016-0406-x (2016).
Shan, Q. et al. Nat. Biotechnol. 31, 686–688 (2013).
Wang, Y. et al. Nat. Biotechnol. 32, 947–951 (2014).
Liang, Z., Zhang, K., Chen, K. & Gao, C. J. Genet. Genomics 41, 63–68 (2014).
Xing, H.L. et al. BMC Plant Biol. 14, 327 (2014).
Kurowska, M. et al. J. Appl. Genet. 52, 371–390 (2011).
Bae, S., Park, J. & Kim, J.S. Bioinformatics 30, 1473–1475 (2014).
Acknowledgements
We acknowledge P. Liu for deep sequencing analysis and T. Li for technical support in flow cytometry. This work was supported by grants from the National Key Research and Development Program of China (2016YFD0101804), the National Transgenic Science and Technology Program (2016ZX08010-002), the Chinese Academy of Sciences KFZD-SW-107 and GJHZ1602 and the National Natural Science Foundation of China (31570369).
Author information
Authors and Affiliations
Contributions
Y.Z., Y.W. and C.G. designed the experiments; Y.Z., Y.W., C.L., R.Z. and K.C. performed the experiments; Y.R. advised on wheat tissue culture work; D.W., J.-L.Q., Y.Z. and C.G. wrote the manuscript.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Corresponding author
Ethics declarations
Competing interests
The authors have submitted a patent application (Application Number 201610998842.X) based on the results reported in this paper.
Integrated supplementary information
Supplementary Figure 1 Complete coding sequences of the NLS-APOBEC1-XTEN-n/dCas9-UGI-NLS fusion cistrons optimized in this study.
The NLSs are written in lower case. The codon-optimized APOBEC1, XTEN linker and UGI are highlighted in green, blue and brown, respectively. The codon-optimized nCas9/dCas9 is shown in bold. The codon marked in yellow indicates the D10A substitution in the HNH domain of Cas9. The H840A substitution in RuvC domain of Cas9 is shown in purple.
Supplementary Figure 2 Analysis of nCas9-PBE (dCas9-PBE) mediated BFP to GFP conversion in wheat protoplasts by flow cytometry.
Four representative fields of protoplasts are shown. Protoplasts were transformed with the following DNA constructs (from left to right): (i) pnCas9-PBE, pUbi-BFPm and pTaU6-BFP-sgRNA, (ii) pdCas9-PBE, pUbi-BFPm and pTaU6-BFP-sgRNA, (iii) pUbi-BFPm and pTaU6-BFP-sgRNA, (iv) a positive control using pUbi-GFP. Scale bars, 800 μm.
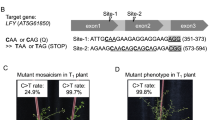
Supplementary Figure 3 Genotypes of 12 representative OsCDC48 point mutants identified by Sanger sequencing.
(a) Schematic of the T-DNA expression vector pH-n/dCas9-PBE for targeting OsCDC48 gene. The coding sequence of hygromycin (Hyg) is driven by 2x35S promoter. (b) Genotypes of twelve representative OsCDC48 point mutants (T0-1 to T0-12) identified by Sanger sequencing. The four C bases shown in red in the wild type (WT) sequence are situated in the deamination window. The T bases marked in red in the 12 mutant sequences are the results of C to T conversion by nCas9-PBE. (c) Sanger sequencing chromatograms for the two homozygous mutant lines (T0-18 and T0-34). The T base resulting from the editing was marked in red. The primers used for Sanger sequencing are listed in Supplementary Table 5.
Supplementary Figure 4 Identification and analysis of the wheat plantlets with targeted C to T conversions by nCas9-PBE.
(a) Sequence of the target site (protospacer) of exon 5 of TaLOX2. The C bases in the deamination window are highlighted in red. The PAM sequence is shown in bold font and the SalI restriction site is underlined. (b) The results of T7E1 and PCR-RE assays analyzing 12 T0 plantlets (T0-1 to T0-12). Two mutants (T0-3 and T0-7) were identified by both T7E1 (top panel) and PCR-RE (mediated by SalI digestion, middle panel) assays. Both assays and Sanger sequencing indicated that T0-3 and T0-7 were heterozygous. WT/D and WT/U indicate the genomic DNA amplicons of wild type (WT) control with or without the digestion by T7E1 or SalI. The bands marked by red arrowheads are diagnostic of positive base editing. The lower panel shows the WT sequence of the target site and its counterpart in the two mutant plantlets (T0-3 and T0-7). The three C bases (displayed in red) in the WT sequence were all converted to T in T0-3, but only one of them was mutated to T in T0-7. The sequences were determined by Sanger sequencing.
Supplementary Figure 5 Constructs used for base editing of TaLOX2 and detection of transgene integration in the resultant T0 mutants.
(a) Diagram of the pnCas9-PBE and pTaU6-LOX2-S1-sgRNA vectors used for base editing of TaLOX2. The positions of the six primer sets (F1/R1, F2/R2, F3/R3, F4/R4, F5/R5 and F6/R6) used for detecting transgene integration are shown. (b) Outcome of the tests for transgene integration using six primer sets for two mutant plants (T0-3 and T0-7). None of the six primer sets yielded the expected PCR amplicon in T0-3 and T0-7, indicating that they were transgene free. The negative control for the tests was performed using the genomic DNA extracted from wild type wheat plants (cv Bobwhite). The positive control for the tests was conducted with the plasmid DNA of pnCas9-PBE or pTaU6-LOX2-S1-sgRNA, with the anticipated products being indicated by red arrowheads.
Supplementary Figure 6 The spectrum of point mutations created using nCas9-PBE in the CENP-A targeting domain (CATD) of ZmCENH3.
In wild type ZmCENH3, this leucine residue is situated in the middle of an alanine-leucine-leucine (ALL, residue 109 to 111) segment. Among the amplicons of the ZmCENH3 target site amplified from protoplasts transformed with nCas9-PBE, we identified reads with A109V, L110F and/or L111F substitutions. We found seven different types of substitutions in ALL, namely single residue substitutions (AFL, VLL and ALF), double substitutions (AFF, VFL and VLF) and triple substitutions (VFF). The CATD regions of Arabidopsis CENH3 (AtCENH3) and barley CENH3 (HvβCENH3) were included in this figure to facilitate comparison. The leucine residue marked in red has previously been shown required for the proper function of AtCENH3 and HvβCENH3. Substitution of the color labeled leucine and alanine residues in AtCENH3 have been found to result in haploid induction. The GenBank accession numbers for ZmCENH3, AtCENH3 and HvβCENH3 are AF519807, AF465800 and JF419329, respectively.
Supplementary Figure 7 Base editing of ZmCENH3 in transgenic maize plants.
(a) Target site (protospacer) sequence of the sgRNA in the exon 3 of ZmCENH3. The C bases in the deamination window are highlighted in red and numbered. The PAM sequence is shown in bold font. (b) Diagram of the T-DNA transformation vector pB-nCas9-PBE for editing ZmCENH3. The coding sequence of Bar (encoding phosphinotricin acetyl transferase) is driven by 2×35S promoter. (c) Sanger sequencing results for heterozygous and homozygous mutants with C7 to T7 substitution. The T base produced by base editing is labeled in red.
Supplementary information
Supplementary Text and Figures
Supplementary Figure 1–7, and Supplementary Tables 1–5 (PDF 3243 kb)
Rights and permissions
About this article
Cite this article
Zong, Y., Wang, Y., Li, C. et al. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat Biotechnol 35, 438–440 (2017). https://doi.org/10.1038/nbt.3811
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.3811
This article is cited by
-
Precise fine-turning of GhTFL1 by base editing tools defines ideal cotton plant architecture
Genome Biology (2024)
-
A mediator of OsbZIP46 deactivation and degradation negatively regulates seed dormancy in rice
Nature Communications (2024)
-
Precise integration of large DNA sequences in plant genomes using PrimeRoot editors
Nature Biotechnology (2024)
-
Genome editing based trait improvement in crops: current perspective, challenges and opportunities
The Nucleus (2024)
-
Fusion of a rice endogenous N-methylpurine DNA glycosylase to a plant adenine base transition editor ABE8e enables A-to-K base editing in rice plants
aBIOTECH (2024)