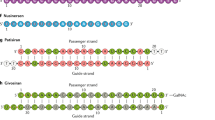
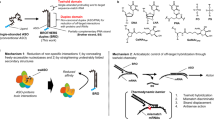
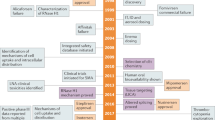
Abstract
After nearly 40 years of development, oligonucleotide therapeutics are nearing meaningful clinical productivity. One of the key advantages of oligonucleotide drugs is that their delivery and potency are derived primarily from the chemical structure of the oligonucleotide whereas their target is defined by the base sequence. Thus, as oligonucleotides with a particular chemical design show appropriate distribution and safety profiles for clinical gene silencing in a particular tissue, this will open the door to the rapid development of additional drugs targeting other disease-associated genes in the same tissue. To achieve clinical productivity, the chemical architecture of the oligonucleotide needs to be optimized with a combination of sugar, backbone, nucleobase, and 3′- and 5′-terminal modifications. A portfolio of chemistries can be used to confer drug-like properties onto the oligonucleotide as a whole, with minor chemical changes often translating into major improvements in clinical efficacy. One outstanding challenge in oligonucleotide chemical development is the optimization of chemical architectures to ensure long-term safety. There are multiple designs that enable effective targeting of the liver, but a second challenge is to develop architectures that enable robust clinical efficacy in additional tissues.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Cohen, J.S. Informational drugs: a new concept in pharmacology. Antisense Res. Dev. 1, 191–193 (1991).
Burnett, J.C. & Rossi, J.J. RNA-based therapeutics: current progress and future prospects. Chem. Biol. 19, 60–71 (2012).
Yamamoto, T., Nakatani, M., Narukawa, K. & Obika, S. Antisense drug discovery and development. Future Med. Chem. 3, 339–365 (2011).
Lindow, M. & Kauppinen, S. Discovering the first microRNA-targeted drug. J. Cell Biol. 199, 407–412 (2012).
Keefe, A.D., Pai, S. & Ellington, A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 9, 537–550 (2010).
Sullenger, B.A. & Nair, S. From the RNA world to the clinic. Science 352, 1417–1420 (2016).
Koch, T., Shim, I., Lindow, M., Ørum, H. & Bohr, H.G. Quantum mechanical studies of DNA and LNA. Nucleic Acid Ther. 24, 139–148 (2014).
Deleavey, G.F. & Damha, M.J. Designing chemically modified oligonucleotides for targeted gene silencing. Chem. Biol. 19, 937–954 (2012).
Sharma, V.K. & Watts, J.K. Oligonucleotide therapeutics: chemistry, delivery and clinical progress. Future Med. Chem. 7, 2221–2242 (2015).
Wan, W.B. & Seth, P.P. The medicinal chemistry of therapeutic oligonucleotides. J. Med. Chem. 59, 9645–9667 (2016).
Ito, K.R. & Obika, S. in Comprehensive Medicinal Chemistry 3rd edn. (eds. Chackalamannil, S. et al.) http://dx.doi.org/10.1016/b978-0-12-409547-2.12420-5 (Elsevier, in the press).
Stephenson, M.L. & Zamecnik, P.C. Inhibition of Rous sarcoma viral RNA translation by a specific oligodeoxyribonucleotide. Proc. Natl. Acad. Sci. USA 75, 285–288 (1978).
Zamecnik, P.C. & Stephenson, M.L. Inhibition of Rous sarcoma virus replication and cell transformation by a specific oligodeoxynucleotide. Proc. Natl. Acad. Sci. USA 75, 280–284 (1978).
Bennett, C.F. & Swayze, E.E. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol. 50, 259–293 (2010).
Eckstein, F. Phosphorothioates, essential components of therapeutic oligonucleotides. Nucleic Acid Ther. 24, 374–387 (2014).
Fluiter, K. Antisense Drug Technology: Principles, Strategies, and Applications. Edited by Stanley T. Crooke. ChemMedChem 4, 879 (2009).
Geary, R.S., Norris, D., Yu, R. & Bennett, C.F. Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv. Drug Deliv. Rev. 87, 46–51 (2015).
Koller, E. et al. Mechanisms of single-stranded phosphorothioate modified antisense oligonucleotide accumulation in hepatocytes. Nucleic Acids Res. 39, 4795–4807 (2011).
Cummins, L.L. et al. Characterization of fully 2′-modified oligoribonucleotide hetero- and homoduplex hybridization and nuclease sensitivity. Nucleic Acids Res. 23, 2019–2024 (1995).
Monia, B.P., Johnston, J.F., Sasmor, H. & Cummins, L.L. Nuclease resistance and antisense activity of modified oligonucleotides targeted to Ha-ras. J. Biol. Chem. 271, 14533–14540 (1996).
Choung, S., Kim, Y.J., Kim, S., Park, H.O. & Choi, Y.C. Chemical modification of siRNAs to improve serum stability without loss of efficacy. Biochem. Biophys. Res. Commun. 342, 919–927 (2006).
Robbins, M. et al. 2′-O-methyl-modified RNAs act as TLR7 antagonists. Mol. Ther. 15, 1663–1669 (2007).
Manoharan, M. 2′-carbohydrate modifications in antisense oligonucleotide therapy: importance of conformation, configuration and conjugation. Biochim. Biophys. Acta 1489, 117–130 (1999).
Freier, S.M. & Altmann, K.-H. The ups and downs of nucleic acid duplex stability: structure-stability studies on chemically-modified DNA:RNA duplexes. Nucleic Acids Res. 25, 4429–4443 (1997).
Mangos, M.M. & Damha, M.J. Flexible and frozen sugar-modified nucleic acids--modulation of biological activity through furanose ring dynamics in the antisense strand. Curr. Top. Med. Chem. 2, 1147–1171 (2002).
Prakash, T.P. An overview of sugar-modified oligonucleotides for antisense therapeutics. Chem. Biodivers. 8, 1616–1641 (2011).
Egli, M. et al. Probing the influence of stereoelectronic effects on the biophysical properties of oligonucleotides: comprehensive analysis of the RNA affinity, nuclease resistance, and crystal structure of ten 2′-O-ribonucleic acid modifications. Biochemistry 44, 9045–9057 (2005).
Martin, P. A New access to 2′-O-alkylated ribonucleosides and properties of 2′-O-alkylated oligoribonucleotides. Helv. Chim. Acta 78, 486–504 (1995).
Kool, E.T. Preorganization of DNA: Design principles for improving nucleic acid recognition by synthetic oligonucleotides. Chem. Rev. 97, 1473–1488 (1997).
Owczarzy, R., You, Y., Groth, C.L. & Tataurov, A.V. Stability and mismatch discrimination of locked nucleic acid-DNA duplexes. Biochemistry 50, 9352–9367 (2011).
Koshkin, A.A. et al. LNA (locked nucleic acids): synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Tetrahedron 54, 3607–3630 (1998).
Obika, S. et al. Stability and structural features of the duplexes containing nucleoside analogues with a fixed N-type conformation, 2′-O,4′-C-methyleneribonucleosides. Tetrahedr. Lett. 39, 5401–5404 (1998).
Singh, S.K., Nielsen, P., Koshkin, A.A. & Wengel, J. LNA (locked nucleic acids): synthesis and high-affinity nucleic acid recognition. Chem. Commun. (Camb.) 1998 455–456 (1998).
Watts, J.K. Locked nucleic acid: tighter is different. Chem. Commun. (Camb.) 49, 5618–5620 (2013).
Ittig, D., Liu, S., Renneberg, D., Schümperli, D. & Leumann, C.J. Nuclear antisense effects in cyclophilin A pre-mRNA splicing by oligonucleotides: a comparison of tricyclo-DNA with LNA. Nucleic Acids Res. 32, 346–353 (2004).
Goyenvalle, A. et al. Functional correction in mouse models of muscular dystrophy using exon-skipping tricyclo-DNA oligomers. Nat. Med. 21, 270–275 (2015).
Jaschinski, F. et al. Next generation antisense oligonucleotides targeting TGF-β. Presented at the 9th annual meeting of the Oligonucleotide Therapeutics Society (Naples, Italy, October 6–8, 2013).
Pedersen, L., Hagedorn, P.H., Lindholm, M.W. & Lindow, M. A kinetic model explains why shorter and less affine enzyme-recruiting oligonucleotides can be more potent. Mol. Ther. Nucleic Acids 3, e149 (2014).
Straarup, E.M. et al. Short locked nucleic acid antisense oligonucleotides potently reduce apolipoprotein B mRNA and serum cholesterol in mice and non-human primates. Nucleic Acids Res. 38, 7100–7111 (2010).
Seth, P.P. et al. Short antisense oligonucleotides with novel 2′-4′ conformationaly restricted nucleoside analogues show improved potency without increased toxicity in animals. J. Med. Chem. 52, 10–13 (2009).
Swayze, E.E. et al. Antisense oligonucleotides containing locked nucleic acid improve potency but cause significant hepatotoxicity in animals. Nucleic Acids Res. 35, 687–700 (2007).
Kamola, P.J. et al. In silico and in vitro evaluation of exonic and intronic off-target effects form a critical element of therapeutic ASO gapmer optimization. Nucleic Acids Res. 43, 8638–8650 (2015).
Burel, S.A. et al. Hepatotoxicity of high affinity gapmer antisense oligonucleotides is mediated by RNase H1 dependent promiscuous reduction of very long pre-mRNA transcripts. Nucleic Acids Res. 44, 2093–2109 (2016).
Kasuya, T. et al. Ribonuclease H1-dependent hepatotoxicity caused by locked nucleic acid-modified gapmer antisense oligonucleotides. Sci. Rep. 6, 30377 (2016).
Østergaard, M.E. et al. Allele-selective inhibition of mutant huntingtin with 2-thio- and C5-triazolylphenyl-deoxythymidine-modified antisense oligonucleotides. Nucleic Acid Ther. 25, 266–274 (2015).
Southwell, A.L. et al. In vivo evaluation of candidate allele-specific mutant huntingtin gene silencing antisense oligonucleotides. Mol. Ther. 22, 2093–2106 (2014).
Østergaard, M.E. et al. Rational design of antisense oligonucleotides targeting single nucleotide polymorphisms for potent and allele selective suppression of mutant Huntingtin in the CNS. Nucleic Acids Res. 41, 9634–9650 (2013).
Damha, M.J. et al. Hybrids of RNA and arabinonucleic acids (ANA and 2′F-ANA) are substrates of Ribonuclease H. J. Am. Chem. Soc. 120, 12976–12977 (1998).
Watts, J.K. & Damha, M.J. 2′F-arabinonucleic acids (2′F-ANA)—history, properties, and new frontiers. Can. J. Chem. 86, 641–656 (2008).
Mangos, M.M. et al. Efficient RNase H-directed cleavage of RNA promoted by antisense DNA or 2′F-ANA constructs containing acyclic nucleotide inserts. J. Am. Chem. Soc. 125, 654–661 (2003).
Min, K.-L., Viazovkina, E., Galarneau, A., Parniak, M.A. & Damha, M.J. Oligonucleotides comprised of alternating 2′-deoxy-2′-fluoro-β-D-arabinonucleosides and D-2′-deoxyribonucleosides (2′F-ANA/DNA 'altimers') induce efficient RNA cleavage mediated by RNase H. Bioorg. Med. Chem. Lett. 12, 2651–2654 (2002).
Mendell, J.R. et al. Eteplirsen for the treatment of Duchenne muscular dystrophy. Ann. Neurol. 74, 637–647 (2013).
Chiriboga, C.A. et al. Results from a phase 1 study of nusinersen (ISIS-SMN(Rx)) in children with spinal muscular atrophy. Neurology 86, 890–897 (2016).
Garber, K. Big win possible for Ionis/Biogen antisense drug in muscular atrophy. Nat. Biotechnol. 34, 1002–1003 (2016).
Li, Z. & Rana, T.M. Therapeutic targeting of microRNAs: current status and future challenges. Nat. Rev. Drug Discov. 13, 622–638 (2014).
Lennox, K.A. & Behlke, M.A. A direct comparison of anti-microRNA oligonucleotide potency. Pharm. Res. 27, 1788–1799 (2010).
Gebert, L.F. et al. Miravirsen (SPC3649) can inhibit the biogenesis of miR-122. Nucleic Acids Res. 42, 609–621 (2014).
Obad, S. et al. Silencing of microRNA families by seed-targeting tiny LNAs. Nat. Genet. 43, 371–378 (2011).
Zanardi, T.A. et al. Pharmacodynamics and subchronic toxicity in mice and monkeys of ISIS 388626, a second-generation antisense oligonucleotide that targets human sodium glucose cotransporter 2. J. Pharmacol. Exp. Ther. 343, 489–496 (2012).
Subramanian, R.R. et al. Enhancing antisense efficacy with multimers and multi-targeting oligonucleotides (MTOs) using cleavable linkers. Nucleic Acids Res. 43, 9123–9132 (2015).
Bhagat, L. et al. Novel oligonucleotides containing two 3′-ends complementary to target mRNA show optimal gene-silencing activity. J. Med. Chem. 54, 3027–3036 (2011).
Boczkowska, M., Guga, P. & Stec, W.J. Stereodefined phosphorothioate analogues of DNA: relative thermodynamic stability of the model PS-DNA/DNA and PS-DNA/RNA complexes. Biochemistry 41, 12483–12487 (2002).
Koziolkiewicz, M., Krakowiak, A., Kwinkowski, M., Boczkowska, M. & Stec, W.J. Stereodifferentiation—the effect of P chirality of oligo(nucleoside phosphorothioates) on the activity of bacterial RNase H. Nucleic Acids Res. 23, 5000–5005 (1995).
Stec, W.J. et al. Stereodependent inhibition of plasminogen activator inhibitor type 1 by phosphorothioate oligonucleotides: proof of sequence specificity in cell culture and in vivo rat experiments. Antisense Nucleic Acid Drug Dev. 7, 567–573 (1997).
Gagnon, K.T. & Watts, J.K. Meeting report: 10th annual meeting of the Oligonucleotide Therapeutics Society. Nucl. Acid Ther. 24, 428–434 (2014).
Fire, A. et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811 (1998).
Elbashir, S.M. et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494–498 (2001).
DeVincenzo, J. et al. A randomized, double-blind, placebo-controlled study of an RNAi-based therapy directed against respiratory syncytial virus. Proc. Natl. Acad. Sci. USA 107, 8800–8805 (2010).
Kanasty, R., Dorkin, J.R., Vegas, A. & Anderson, D. Delivery materials for siRNA therapeutics. Nat. Mater. 12, 967–977 (2013).
Corey, D.R. RNA learns from antisense. Nat. Chem. Biol. 3, 8–11 (2007).
Layzer, J.M. et al. In vivo activity of nuclease-resistant siRNAs. RNA 10, 766–771 (2004).
Watts, J.K., Deleavey, G.F. & Damha, M.J. Chemically modified siRNA: tools and applications. Drug Discov. Today 13, 842–855 (2008).
Jackson, A.L. et al. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA 12, 1197–1205 (2006).
Bartlett, D.W. & Davis, M.E. Effect of siRNA nuclease stability on the in vitro and in vivo kinetics of siRNA-mediated gene silencing. Biotechnol. Bioeng. 97, 909–921 (2007).
Allerson, C.R. et al. Fully 2′-modified oligonucleotide duplexes with improved in vitro potency and stability compared to unmodified small interfering RNA. J. Med. Chem. 48, 901–904 (2005).
Prakash, T.P. et al. Positional effect of chemical modifications on short interference RNA activity in mammalian cells. J. Med. Chem. 48, 4247–4253 (2005).
Morrissey, D.V. et al. Activity of stabilized short interfering RNA in a mouse model of hepatitis B virus replication. Hepatology 41, 1349–1356 (2005).
Deleavey, G.F. et al. Synergistic effects between analogs of DNA and RNA improve the potency of siRNA-mediated gene silencing. Nucleic Acids Res. 38, 4547–4557 (2010).
Deleavey, G.F., Watts, J.K. & Damha, M.J. Chemical modification of siRNA. Curr. Protoc. Nucleic Acid Chem. 16, 16.3 (2009).
Dar, S.A., Thakur, A., Qureshi, A. & Kumar, M. siRNAmod: A database of experimentally validated chemically modified siRNAs. Sci. Rep. 6, 20031 (2016).
Snead, N.M., Escamilla-Powers, J.R., Rossi, J.J. & McCaffrey, A.P. 5′ unlocked nucleic acid modification improves siRNA targeting. Mol. Ther. Nucleic Acids 2, e103 (2013).
Soutschek, J. et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature 432, 173–178 (2004).
Czauderna, F. et al. Structural variations and stabilising modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res. 31, 2705–2716 (2003).
Manoharan, M. et al. Unique gene-silencing and structural properties of 2′-fluoro-modified siRNAs. Angew. Chem. Int. Ed. Engl. 50, 2284–2288 (2011).
Cuellar, T.L. et al. Systematic evaluation of antibody-mediated siRNA delivery using an industrial platform of THIOMAB–siRNA conjugates. Nucleic Acids Res. 43, 1189–1203 (2015)<>.
Matranga, C., Tomari, Y., Shin, C., Bartel, D.P. & Zamore, P.D. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 123, 607–620 (2005).
Schirle, N.T., Sheu-Gruttadauria, J. & MacRae, I.J. Structural basis for microRNA targeting. Science 346, 608–613 (2014).
Addepalli, H. et al. Modulation of thermal stability can enhance the potency of siRNA. Nucleic Acids Res. 38, 7320–7331 (2010).
Rajeev, K.G. et al. RNAi agents, compositions and methods of use thereof for treating transthyretin (TTR) associated diseases. US patent 9,399,775 (2016).
Khvorova, A., Reynolds, A. & Jayasena, S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell 115, 209–216 (2003).
Salomon, W.E., Jolly, S.M., Moore, M.J., Zamore, P.D. & Serebrov, V. Single-molecule imaging reveals that Argonaute reshapes the binding properties of its nucleic acid guides. Cell 162, 84–95 (2015).
Schirle, N.T. et al. Structural analysis of human Argonaute-2 bound to a modified siRNA guide. J. Am. Chem. Soc. 138, 8694–8697 (2016).
Kel'in, A.V. et al. Structural basis of duplex thermodynamic stability and enhanced nuclease resistance of 5′-C-methyl pyrimidine-modified oligonucleotides. J. Org. Chem. 81, 2261–2279 (2016).
Martinez, J., Patkaniowska, A., Urlaub, H., Lührmann, R. & Tuschl, T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110, 563–574 (2002).
Frank, F., Sonenberg, N. & Nagar, B. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature 465, 818–822 (2010).
Ma, J.B. et al. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature 434, 666–670 (2005).
Weitzer, S. & Martinez, J. The human RNA kinase hClp1 is active on 3′ transfer RNA exons and short interfering RNAs. Nature 447, 222–226 (2007).
Chen, P.Y. et al. Strand-specific 5′-O-methylation of siRNA duplexes controls guide strand selection and targeting specificity. RNA 14, 263–274 (2008).
Kenski, D.M. et al. siRNA-optimized modifications for enhanced in vivo activity. Mol. Ther. Nucleic Acids 1, e5 (2012).
Parmar, R. et al. 5′-(E)-vinylphosphonate: a stable phosphate mimic can improve the RNAi Activity of siRNA-GalNAc conjugates. ChemBioChem 17, 985–989 (2016).
Yu, D. et al. Single-stranded RNAs use RNAi to potently and allele-selectively inhibit mutant huntingtin expression. Cell 150, 895–908 (2012).
Lima, W.F. et al. Single-stranded siRNAs activate RNAi in animals. Cell 150, 883–894 (2012).
Prakash, T.P. et al. Identification of metabolically stable 5-phospha 5(-phosphate analogs that support single-stranded siRNA activity. Nucleic Acids Res. 43, 2993–3011 (2015).
Davis, M.E. et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature 464, 1067–1070 (2010).
Coelho, T. et al. Safety and efficacy of RNAi therapy for transthyretin amyloidosis. N. Engl. J. Med. 369, 819–829 (2013).
Juliano, R.L. The delivery of therapeutic oligonucleotides. Nucleic Acids Res. 44, 6518–6548 (2016).
Nair, J.K. et al. Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J. Am. Chem. Soc. 136, 16958–16961 (2014).
Prakash, T.P. et al. Synergistic effect of phosphorothioate, 5′-vinylphosphonate and GalNAc modifications for enhancing activity of synthetic siRNA. Bioorg. Med. Chem. Lett. 26, 2817–2820 (2016).
Mäkilä, J. et al. Synthesis of multi-galactose-conjugated 2′-O-methyl oligoribonucleotides and their in vivo imaging with positron emission tomography. Bioorg. Med. Chem. 22, 6806–6813 (2014).
Fitzgerald, K. et al. A highly durable RNAi therapeutic inhibitor of PCSK9. N. Engl. J. Med. 376, 41–51 (2017).
Shen, W., Liang, X.H., Sun, H. & Crooke, S.T. 2′-fluoro-modified phosphorothioate oligonucleotide can cause rapid degradation of P54nrb and PSF. Nucleic Acids Res. 43, 4569–4578 (2015).
Janas, M.M. et al. Impact of oligonucleotide structure, chemistry, and delivery method on in vitro cytotoxicity. Nucleic Acid Ther. http://dx.doi.org/10.1089/nat.2016/0639 (2016).
Matsuda, S. et al. iRNA conjugates carrying sequentially assembled trivalent N-acetylgalactosamine linked through nucleosides elicit robust gene silencing in vivo in hepatocytes. ACS Chem. Biol. 10, 1181–1187 (2015).
Rajeev, K.G. et al. Hepatocyte-specific delivery of siRNAs conjugated to novel non-nucleosidic trivalent N-acetylgalactosamine elicits robust gene silencing in vivo. ChemBioChem 16, 903–908 (2015).
Sehgal, A. et al. An RNAi therapeutic targeting antithrombin to rebalance the coagulation system and promote hemostasis in hemophilia. Nat. Med. 21, 492–497 (2015).
Prakash, T.P. et al. Targeted delivery of antisense oligonucleotides to hepatocytes using triantennary N-acetyl galactosamine improves potency 10-fold in mice. Nucleic Acids Res. 42, 8796–8807 (2014).
Yu, R.Z. et al. Disposition and pharmacology of a GalNAc3-conjugated ASO targeting human lipoprotein (a) in mice. Mol. Ther. Nucleic Acids 5, e317 (2016).
Yu, R.Z. et al. Disposition and pharmacokinetics of a GalNAc3-conjugated antisense oligonucleotide targeting human lipoprotein (a) in monkeys. Nucleic Acid Ther. 26, 372–380 (2016).
Wolfrum, C. et al. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat. Biotechnol. 25, 1149–1157 (2007).
Byrne, M. et al. Novel hydrophobically modified asymmetric RNAi compounds (sd-rxRNA) demonstrate robust efficacy in the eye. J. Ocul. Pharmacol. Ther. 29, 855–864 (2013).
Alterman, J.F. et al. Hydrophobically modified siRNAs silence huntingtin mRNA in primary neurons and mouse brain. Mol. Ther. Nucleic Acids 4, e266 (2015).
Nikan, M. et al. Docosahexaenoic acid conjugation enhances distribution and safety of siRNA upon local administration in mouse brain. Mol. Ther. Nucleic Acids 5, e344 (2016).
Thompson, J.D. et al. Toxicological and pharmacokinetic properties of chemically modified siRNAs targeting p53 RNA following intravenous administration. Nucleic Acid Ther. 22, 255–264 (2012).
Stein, C.A. et al. G3139, an anti-Bcl-2 antisense oligomer that binds heparin-binding growth factors and collagen I, alters in vitro endothelial cell growth and tubular morphogenesis. Clin. Cancer Res. 15, 2797–2807 (2009).
Khorkova, O. & Wahlestedt, C. Oligonucleotide therapies for disorders of the nervous system. Nat. Biotechnol. 35, 250–264 (2017).
Lennox, K.A. & Behlke, M.A. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res. 44, 863–877 (2016).
Gagnon, K.T. et al. Allele-selective inhibition of mutant huntingtin expression with antisense oligonucleotides targeting the expanded CAG repeat. Biochemistry 49, 10166–10178 (2010).
Stalder, L. et al. The rough endoplasmatic reticulum is a central nucleation site of siRNA-mediated RNA silencing. EMBO J. 32, 1115–1127 (2013).
Pei, Y. et al. Quantitative evaluation of siRNA delivery in vivo. RNA 16, 2553–2563 (2010).
Schwartz, J.C. et al. Antisense transcripts are targets for activating small RNAs. Nat. Struct. Mol. Biol. 15, 842–848 (2008).
Modarresi, F. et al. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat. Biotechnol. 30, 453–459 (2012).
Woo, C. et al. Poster Presented at the Harvard Epigenetics Symposium, Boston, MA, October 22, 2015.
Li, L., Matsui, M. & Corey, D.R. Activating frataxin expression by repeat-targeted nucleic acids. Nat. Commun. 7, 10606 (2016).
Nomakuchi, T.T., Rigo, F., Aznarez, I. & Krainer, A.R. Antisense oligonucleotide-directed inhibition of nonsense-mediated mRNA decay. Nat. Biotechnol. 34, 164–166 (2016).
Liang, X.H. et al. Translation efficiency of mRNAs is increased by antisense oligonucleotides targeting upstream open reading frames. Nat. Biotechnol. 34, 875–880 (2016).
Jiang, K. Biotech comes to its 'antisenses' after hard-won drug approval. Spoonful of Medicine http://blogs.nature.com/spoonful/2013/02/biotech-comes-to-its-antisenses-after-hard-won-drug-approval.html (19 February 2013).
World Health Organzation. Recommended international nonproprietary names for pharmaceutical substances (INN): list 76. WHO Drug Information 30, 477–544 (2016).
Khvorova, A. Oligonucleotide therapeutics—a new class of cholesterol-lowering drugs. N. Engl. J. Med. 376, 4–7 (2017).
Acknowledgements
We thank M. Osborn for the oligonucleotide image in Figure 1. We would like to acknowledge the oligonucleotide research community who contributed to the evolution of this field, including those whose excellent work we could not mention for lack of space. We thank D. Conte for significant help with manuscript preparation. This work was supported by the RNA Therapeutics Institute of University of Massachusetts Medical School, the CHDI Foundation, and US National Institutes of Health grants R01GM1088030181, R01HD086111, and UH3TR000888 to A.K.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
A.K. owns stock in RXi Pharmaceuticals.
Rights and permissions
About this article
Cite this article
Khvorova, A., Watts, J. The chemical evolution of oligonucleotide therapies of clinical utility. Nat Biotechnol 35, 238–248 (2017). https://doi.org/10.1038/nbt.3765
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.3765
This article is cited by
-
Enhanced bacterial cancer therapy delivering therapeutic RNA interference of c-Myc
Cell & Bioscience (2024)
-
Chemically modified microRNA delivery via DNA tetrahedral frameworks for dental pulp regeneration
Journal of Nanobiotechnology (2024)
-
Novel STAT3 oligonucleotide compounds suppress tumor growth and overcome the acquired resistance to sorafenib in hepatocellular carcinoma
Acta Pharmacologica Sinica (2024)
-
RNAi-based drug design: considerations and future directions
Nature Reviews Drug Discovery (2024)
-
Alternative therapeutic strategies to treat antibiotic-resistant pathogens
Nature Reviews Microbiology (2024)