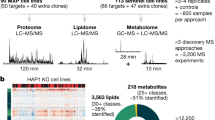
Abstract
Mitochondrial dysfunction is associated with many human diseases, including cancer and neurodegeneration, that are often linked to proteins and pathways that are not well-characterized. To begin defining the functions of such poorly characterized proteins, we used mass spectrometry to map the proteomes, lipidomes, and metabolomes of 174 yeast strains, each lacking a single gene related to mitochondrial biology. 144 of these genes have human homologs, 60 of which are associated with disease and 39 of which are uncharacterized. We present a multi-omic data analysis and visualization tool that we use to find covariance networks that can predict molecular functions, correlations between profiles of related gene deletions, gene-specific perturbations that reflect protein functions, and a global respiration deficiency response. Using this multi-omic approach, we link seven proteins including Hfd1p and its human homolog ALDH3A1 to mitochondrial coenzyme Q (CoQ) biosynthesis, an essential pathway disrupted in many human diseases. This Resource should provide molecular insights into mitochondrial protein functions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Hebert, A.S. et al. The one hour yeast proteome. Mol. Cell. Proteomics 13, 339–347 (2014).
Richards, A.L. et al. One-hour proteome analysis in yeast. Nat. Protoc. 10, 701–714 (2015).
Peterson, A.C. et al. Development of a GC/Quadrupole-Orbitrap mass spectrometer, part I: design and characterization. Anal. Chem. 86, 10036–10043 (2014).
Ishii, N. et al. Multiple high-throughput analyses monitor the response of E. coli to perturbations. Science 316, 593–597 (2007).
Buescher, J.M. et al. Global network reorganization during dynamic adaptations of Bacillus subtilis metabolism. Science 335, 1099–1103 (2012).
Williams, E.G. et al. Systems proteomics of liver mitochondria function. Science 352, aad0189 (2016).
Chick, J.M. et al. Defining the consequences of genetic variation on a proteome-wide scale. Nature 534, 500–505 (2016).
Nunnari, J. & Suomalainen, A. Mitochondria: in sickness and in health. Cell 148, 1145–1159 (2012).
Koopman, W.J., Willems, P.H. & Smeitink, J.A. Monogenic mitochondrial disorders. N. Engl. J. Med. 366, 1132–1141 (2012).
Vafai, S.B. & Mootha, V.K. Mitochondrial disorders as windows into an ancient organelle. Nature 491, 374–383 (2012).
Pagliarini, D.J. et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell 134, 112–123 (2008).
Calvo, S.E., Clauser, K.R. & Mootha, V.K. MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 44, D1, D1251–D1257 (2016).
Sickmann, A. et al. The proteome of Saccharomyces cerevisiae mitochondria. Proc. Natl. Acad. Sci. USA 100, 13207–13212 (2003).
Green, E.D. & Guyer, M.S. Charting a course for genomic medicine from base pairs to bedside. Nature 470, 204–213 (2011).
Pagliarini, D.J. & Rutter, J. Hallmarks of a new era in mitochondrial biochemistry. Genes Dev. 27, 2615–2627 (2013).
Floyd, B.J. et al. Mitochondrial protein interaction mapping identifies new regulators of respiratory chain function. Mol. Cell 63, 621–632 (2016).
Quinzii, C.M. & Hirano, M. Coenzyme Q and mitochondrial disease. Dev. Disabil. Res. Rev. 16, 183–188 (2010).
Kalén, A., Appelkvist, E.L. & Dallner, G. Age-related changes in the lipid compositions of rat and human tissues. Lipids 24, 579–584 (1989).
Bentley, R.R.V.G., Springer, C.M., Dialameh, G.H. & Olson, R.E. The origin of the benzoquinone ring of coenzyme Q9 in the rat. Biochem. Biophys. Res. Commun. 5, 443–446 (1961).
Booth, A.N.M.M.S., Robbins, D.J., Emerson, O.H., Jones, F.T. & Deeds, F. Urinary Phenolic Acid Metabolites of Tyrosine. J. Biol. Chem. 235, 2649–2652 (1960).
Pierrel, F. et al. Involvement of mitochondrial ferredoxin and para-aminobenzoic acid in yeast coenzyme Q biosynthesis. Chem. Biol. 17, 449–459 (2010).
Marbois, B. et al. para-Aminobenzoic acid is a precursor in coenzyme Q6 biosynthesis in Saccharomyces cerevisiae. J. Biol. Chem. 285, 27827–27838 (2010).
Nakahara, K. et al. The Sjögren-Larsson syndrome gene encodes a hexadecenal dehydrogenase of the sphingosine 1-phosphate degradation pathway. Mol. Cell 46, 461–471 (2012).
Liu, Z.J. et al. The first structure of an aldehyde dehydrogenase reveals novel interactions between NAD and the Rossmann fold. Nat. Struct. Biol. 4, 317–326 (1997).
Jackson, B. et al. Update on the aldehyde dehydrogenase gene (ALDH) superfamily. Hum. Genomics 5, 283–303 (2011).
De Laurenzi, V. et al. Sjögren-Larsson syndrome is caused by mutations in the fatty aldehyde dehydrogenase gene. Nat. Genet. 12, 52–57 (1996).
Kitamura, T. et al. Substrate specificity, plasma membrane localization, and lipid modification of the aldehyde dehydrogenase ALDH3B1. Biochim. Biophys. Acta 1831, 1395–1401 (2013).
Hughes, T.R. et al. Functional discovery via a compendium of expression profiles. Cell 102, 109–126 (2000).
Kemmeren, P. et al. Large-scale genetic perturbations reveal regulatory networks and an abundance of gene-specific repressors. Cell 157, 740–752 (2014).
Giaever, G. et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418, 387–391 (2002).
Laredj, L.N., Licitra, F. & Puccio, H.M. The molecular genetics of coenzyme Q biosynthesis in health and disease. Biochimie 100, 78–87 (2014).
Tran, U.C. & Clarke, C.F. Endogenous synthesis of coenzyme Q in eukaryotes. Mitochondrion 7 (Suppl.), S62–S71 (2007).
Züchner, S. et al. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat. Genet. 36, 449–451 (2004).
Mourier, A. et al. Mitofusin 2 is required to maintain mitochondrial coenzyme Q levels. J. Cell Biol. 208, 429–442 (2015).
Hebert, A.S. et al. Amine-reactive neutron-encoded labels for highly plexed proteomic quantitation. Mol. Cell. Proteomics 12, 3360–3369 (2013).
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008).
Elias, J.E. & Gygi, S.P. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 (2007).
Cox, J. et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 (2011).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 (1995).
Edgar, R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Larkin, M.A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007).
Letunic, I. & Bork, P. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 39, W475–W478 (2011).
Acknowledgements
We thank members of the Pagliarini and Coon laboratories for helpful discussions. This work was supported by a Searle Scholars Award and NIH grants R01DK098672, R01GM112057, and R01GM115591 (to D.J.P.); NIH grant R35GM118110 (to J.J.C.); NIH Ruth L. Kirschstein NRSA F30AG043282 (to J.A.S.); DOE Great Lakes Bioenergy Research Center (DOE Office of Science BER DE-FC02-07ER64494 to N.W.K. and A.U.); and ACS Analytical Chemistry and Society of Analytical Chemists of Pittsburgh awards (to A.L.R.); NSF Graduate Research Fellowship and NIH T32GM007215 (to M.T.V.); NIH T32DK007665 (to Z.A.K.); and NIH T32HG002760 (to E.A.T.).
Author information
Authors and Affiliations
Contributions
J.A.S., N.W.K., D.J.P., and J.J.C. conceived of the project and its design and wrote the manuscript. J.A.S., A.J., K.P.R., X.G., Z.A.K., and J.S. prepared samples and performed biochemical experiments. E.C.F., A.L.R., M.J.P.R., A.U., P.D.H., X.G., K.J.C., E.A.T., and A.S.H. acquired MS data. J.A.S., N.W.K., M.T.V., H.M., M.S.W., D.J.P., and J.J.C. analyzed data.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Δgene target strain characteristics and respiration culture optimization.
(a) Proteins encoded by the individual genes knocked out of the 174 yeast strains investigated in this study, shown in the context of biological pathways. APS, adenosine-5’-phosphosulfate; CII–CV, oxidative phosphorylation complexes II–V; ER, endoplasmic reticulum; EMC, ER membrane complex; ERMES, ER-mitochondria encounter structure; ETF, electron transfer flavoprotein complex; MAM, mitochondria-associated membrane; MECA, mitochondria-ER-cortex anchor; MICOS, mitochondrial contact site and cristae organizing system; MIM, mitochondrial inner membrane; MOM, mitochondrial outer membrane; mtDNA, mitochondrial DNA; mtRibosome, mitochondrial ribosome; NAD, nicotinamide adenine dinucleotide; PDH, pyruvate dehydrogenase; TCA, tricarboxylic acid cycle; vCLAMP, vacuole and mitochondria patch. The pie charts show the total number of characterized and uncharacterized genes profiled (top); the total number of profiled genes that have human homologs (upper middle); of these genes with human homologs, the number of profiled genes that are also associated with disease (lower middle); and of the uncharacterized genes profiled, the number of genes that have human homologs (bottom). (b) Density of yeast cultures in the respiratory growth condition (mean, n = 3) plotted in strain rank order (left) or against fermentation culture density (mean, n = 3) (right). (c) Optical density at 600 nm (OD600) of yeast cultures (media with 3% [w/v] glycerol and 0.1% [w/v] glucose) indicating time points at which yeast were harvested during fermentation (F1–F3) or respiration (R4–R8). Time point R6 (25 h) was selected for the respiration culture condition of the larger study. (d) Whole-proteome plot of protein abundances at time points R5 and R8. (e) Pairwise whole proteome plot comparisons (as in d) across all eight time points (lower left) and linear regression analysis of each comparison (r2, Pearson correlation coefficients) (upper right).
Supplementary Figure 2 Mass spectrometry analysis metrics and quality assessment.
(a) Proteins, lipids, and metabolites quantified per Δgene strain (mean ± s.d., n = 3). (b) MS experiments conducted per day (top) and phenotypes (molecules) quantified per day (bottom) for proteomics, lipidomics, and metabolomics. (c) Overview of the yeast protein extraction method optimized for this study compared to previous work. (d) Violin plots depicting the range of fold changes in molecule abundance (log2[Δgene/WT]) across all molecule classes and metabolic states. (e) Density plots of the distribution of coefficients of variation (CVs) (%) for each molecule measured in biological triplicate across all mutants and growth conditions. (f) Venn diagrams depicting the average overlap of molecules quantified within individual Δgene strains across fermentation and respiration growth conditions. (g) Average profile overlap between different Δgene strains.
Supplementary Figure 3 Features of protein-lipid-metabolite perturbation profiles.
(a) Heat maps depicting the number of molecules significantly perturbed within each Δgene strain (P < 0.05; two-sided Student’s t-test). (b) Hierarchical clusters of Δgene strains and significantly perturbed molecules (relative abundances compared to WT quantified by MS; P < 0.05; two-sided Student’s t-test). The center column annotates select clusters with significant functional (GO term) enrichments (P < 0.05; Fisher’s exact test followed by Benjamini-Hochberg FDR correction for multiple hypothesis testing). Pie charts indicate proteins in clusters encoded by characterized (gray) or uncharacterized (red) genes.
Supplementary Figure 4 Expanded view of two protein clusters from the respiration Y3K dataset heat map (respiration profiles).
Heat map indicates relative abundance of proteins in Δgene strains compared to WT as quantified by MS. See Supplementary Fig. 3 for the full heat map.
Supplementary Figure 5 Subsets of the Δgene-specific phenotypes identified in this study.
Relative abundances of individual molecules (mean log2[Δgene/WT], n = 3) (x-axes) versus statistical significance (–log10[p-value]; two-sided Student’s t-test) (y-axes) as quantified by MS. The plots shown represent a subset of molecules identified as ‘Δgene-specific phenotypes’ through an unbiased survey of the Y3K dataset (see Fig. 2a). The array here is limited to the most robust outliers (based on both statistical significance and fold-change, see Supplementary Note 2 and Online Methods)—the top 20 upregulated proteins, the top 20 downregulated proteins, the top 10 metabolites, and the top 4 or 5 lipids—excluding ‘knocked out proteins’ (e.g. Fmp52p in the Δfmp52 strain) and excluding a given Δgene strain after it appeared twice on the rank list. Biological hypotheses surrounding gene-phenotype relationship were generated for the starred plots (see Supplementary Fig. 6).
Supplementary Figure 6 Examples of hypotheses that can be generated from a subset of the Δgene-specific phenotypes identified in this study.
Subset of Δgene-specific phenotypes identified in the Y3K dataset. Volcano plots indicate relative molecule abundances (mean log2[Δgene/WT], n = 3) (x-axes) versus statistical significance (–log10[p-value]; two-sided Student’s t-test) (y-axes) as quantified by MS. Hypotheses were developed to describe each Δgene-phenotype relationship reported here.
Supplementary Figure 7 Hfd1p supports production of 4-HB for CoQ biosynthesis.
(a) Relative lipid abundances (mean, n = 3) versus statistical significance (–log10[p-value]; two-sided Student’s t-test) as quantified by MS. (b) Relative abundances of 4-HBz (mean, n = 3) versus statistical significance (–log10[p-value]; two-sided Student’s t-test) across all Δgene strains in the study. (c) Protein domain structures of Hfd1p, highlighting residues involved in catalysis. (d) Serial dilutions of Δhfd1 yeast transformed with plasmids encoding the indicated Hfd1p variants grown on pABA– synthetic solid medias with glucose or glycerol. (e) Relative respiratory growth rates of Δhfd1 yeast transformed with plasmids encoding the indicated Hfd1p variants and grown in pABA– synthetic liquid media. (f) Growth curves showing the respiratory growth of Δhfd1 yeast in pABA– synthetic media with the additives shown. (g) Relative 4-HB abundance in Δhfd1 yeast cultured in pABA– media with the additives shown (mean log2[additive/unsupplemented] ± s.d., n = 3). (h) SDS-PAGE analysis (Coomassie stained gel) of protein fractions from an isolation of MBP-Hfd1p(CΔ25), MBP-ALDH3A1, and MBP-ALDH3A2(CΔ25) (WT and catalytically dead mutant for each). (i) Phylogenetic tree of human ALDH superfamily members and yeast Hfd1p. (j) Density of yeast (upon harvest) cultured in pABA– media ± 4-HB (mean ± s.d., n = 3). (k) Relative abundances of 4-HB, PPHB, and CoQ compared to WT yeast cultured in pABA– media (mean log2[Δgene/WT with no additive] ± s.d., n = 3) as quantified by MS. (l) Whole proteome correlation map for yeast grown in pABA– media ± 4-HB (mean, n = 3). (m) Relative abundances of select proteins as quantified by MS (mean log2[Δgene/WT], n = 3) analysis of yeast cultured in pABA– media ± 4-HB. (n) Serial dilutions of Δhfd1 yeast transformed with plasmids encoding the proteins shown and cultured on solid pABA– synthetic media plates. (o) Enzyme activity of MBP-ALDH3A1 or MBP-ALDH3A2(CΔ25) against 4-HBz (200 μM) or hexadecanal (200 μM) (mean ± s.e.m., n = 3). (p) Table of enzyme kinetic parameters for MBP-Hfd1p(CΔ25), MBP-ALDH3A1, and MBP-ALDH3A2(CΔ25) (mean ± s.e.m., n = 3). (q) Representative enzyme kinetic curves for MBP-ALDH3A1 and MBP-ALDH3A2(CΔ25). *P < 0.05; **P < 0.01; ***P < 0.001 (two-sided Student’s t-test).
Supplementary Figure 8 Identification of respiration deficiency response pathways and potential biomarkers.
(a) Projection of RC and RD strains onto the planes defined by principal component (PC) axes 1 and 2 for separate proteome, metabolome, and lipidome PC analyses. (b) RD versus RC proteome perturbation volcano plots (as in Fig. 3e) showing select functional groups (GO terms) significantly enriched (Bonferroni corrected p-values shown in figure) in either upregulated or downregulated proteins. (c) Box plots depicting median molecule fold changes for RC and RD strains (log2[RD or RC average/WT]) (n = 111 for RC, 41 for RD). Notch indicates 95% c.i. (d) Receiver operating characteristic (ROC) curves for select molecules depicting the false positive rates and true positive rates for prediction of respiration deficiency associated with particular molecule fold changes. AUC, area under the curve.
Supplementary Figure 9 Subtraction of shared responses to reveal deeper biochemical insight.
(a) RDR-abundance adjustment of a representative molecule (Mls1p) by subtraction of the average fold change in abundance (mean log2[Δgene/WT], n = 3) across respiration deficient (RD) strains. This adjustment was only performed within RD strains. (b) Plots comparing relative protein abundances between pairs of Δgene strains. Linear regression analysis of pairs of perturbation profiles before (left) and after (right) RD-abundance adjustment. Green points indicate molecules significantly perturbed in both mutants (|log2(FC)| > 0.7; P < 0.05; two-sided Student’s t-test) prior to RDR-adjustment. (c) Expanded view of highly correlated strains in the respiration proteomes correlation map (see Fig. 3b). (d) Procedure for normalization of the RDR. (e) Re-clustered respiration proteome strain-strain correlation map following RDR-adjustment (also shown in Fig. 3g).
Supplementary Figure 10 Molecular perturbations of yeast lacking yjr120w.
(a) Relative protein abundances (mean log2[Δyjr120w/WT], n = 3) versus statistical significance (–log10[p-value]; two-sided Student’s t-test) as quantified by MS. (b) Relative Atp2p protein abundance (mean log2[Δgene/WT], n = 3) versus statistical significance (–log10[p-value]; two-sided Student’s t-test) across all mutants in the study. (c) Genomic organization of yjr120w and atp2. (d) Serial dilutions of yeast transformed with the indicated plasmids grown on agar plates with glucose (to enable fermentation) or glycerol (to force respiration). (e) Fold changes in mRNA abundances (mean Δgene/WT, n = 3) as quantified by real time polymerase chain reaction (RT-PCR) analysis. Yjr120w mRNA was not detected (n.d.) in WT yeast, so imputation of this missing value was used to calculate the fold increase in yjr120w mRNA shown for the Δatp2 strain. *P < 0.05; **P < 0.01; ***P < 0.001 (two-sided Student’s t-test).
Supplementary Figure 11 Features of multi-omic molecule covariance networks.
(a) Network of all covariant molecules observed in each dataset (|ρ| ≥ 0.58, Bonferroni-adjusted P < 0.001; two-sided Student’s t-test). (b) Regression analysis of pairs of RDR-associated molecules before and after RDR adjustment using Spearman’s rank coefficient (ρ). Points corresponding to RD and RC Δgene strains are indicated. (c) Distribution of calculated Spearman coefficients for all pairwise molecule covariance comparisons (ρ cutoff at ±0.58 used throughout the study is indicated). (d) Distribution of Bonferroni-adjusted p-values from all pairwise molecule comparisons (p-value cutoff at 0.001 used throughout the study is indicated). (e) Bar chart indicating number of protein–protein (P–P), protein–metabolite (P–M), protein–lipid (P–L), metabolite–metabolite (M–M), metabolite–lipid (M–L), and lipid–lipid (L–L) edges in each dataset. (f) Box plots indicating the number of edges per node in the respiration, fermentation, and RDR-adjusted networks. (g) Network of all covariant RDR-associated molecules (|ρ| ≥ 0.58, Bonferroni-adjusted P < 0.001; two-sided Student’s t-test) generated using the respiration (left) and RDR-adjusted (right) datasets. Nodes are highlighted according to GO category. (h) Box plots indicating the molecule covariance network (MCN) specificity coefficient for all nodes involved in mitochondrial translation in both the respiration and RDR-adjusted respiration RDR-associated molecule networks (shown in panel G). (i) Relative protein abundances (mean log2[Δyor020w-a/WT], n = 2) versus statistical significance (–log10[p-value]; two-sided Student’s t-test) as quantified by MS.
Supplementary Figure 12 Molecule covariance networks for uncharacterized proteins.
‘Nearest neighbor’ molecule covariance networks for all uncharacterized proteins observed across the respiration, fermentation, and RDR-adjusted respiration datasets (|ρ| ≥ 0.58, Bonferroni-adjusted P < 0.001; two-sided Student’s t-test). If more than 14 correlated molecules were present in a given covariance network, only the top 14 correlated molecules (nearest neighbors) are displayed.
Supplementary Figure 13 Examples of hypotheses that can be generated from a subset of the molecule covariance network analyses in this study.
Nearest neighbor molecule covariance networks from uncharacterized proteins containing more than four connected nodes were tested for GO term enrichment using a Fisher’s exact test with Benjamini–Hochberg FDR adjustment to account for multiple hypothesis testing. Networks containing four or fewer connected nodes were analyzed manually for functionally related molecules. Based on these MCNA results, biological hypotheses about the functions of the uncharacterized proteins shown were developed.
Supplementary Figure 14 Hypothesized pathways for Aro9p, Aro10p, and Aim18p.
(a) Putative biochemical functions of Aro9p and Aro10p in catabolism of tyrosine and phenylalanine. (b) Predicted functions for Aro9p and Aro10p in the Tyr-to-4-HB-to-CoQ pathway. (c) Protein sequence alignments of Aim18p (S. cerevisiae) and chalcone isomerases (CHI) from Medicago and Arabidopsis highlighting conservation of putative catalytic residues (starred residues). (d) Example of a CHI catalyzed reaction (upper scheme) and the hypothesized pathway of Aim18p action (lower scheme).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–14 and Supplementary Notes 1–7 (PDF 4245 kb)
Supplementary Table 1
Knockout yeast strains. (XLSX 71 kb)
Supplementary Table 2
Profiled biomolecules (XLSX 221 kb)
Supplementary Table 3
Quantitative dataset (XLSX 50846 kb)
Supplementary Table 4
gene-specific phenotypes. (XLSX 52 kb)
Supplementary Table 5
Respiration deficient strains vs respiration competent strains. (XLSX 250 kb)
Supplementary Table 6
Respiration deficient strains versus wild type. (XLSX 88 kb)
Supplementary Table 7
gene-Δgene perturbation profile correlations. (XLSX 235 kb)
Supplementary Table 8
Molecule covariance network analysis results (XLSX 12234 kb)
Supplementary Code
Source Code (ZIP 220660 kb)
Rights and permissions
About this article
Cite this article
Stefely, J., Kwiecien, N., Freiberger, E. et al. Mitochondrial protein functions elucidated by multi-omic mass spectrometry profiling. Nat Biotechnol 34, 1191–1197 (2016). https://doi.org/10.1038/nbt.3683
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.3683
This article is cited by
-
Mitochondrial proteome research: the road ahead
Nature Reviews Molecular Cell Biology (2024)
-
Genetic perturbation of mitochondrial function reveals functional role for specific mitonuclear genes, metabolites, and pathways that regulate lifespan
GeroScience (2023)
-
Proteome effects of genome-wide single gene perturbations
Nature Communications (2022)
-
UbiB proteins regulate cellular CoQ distribution in Saccharomyces cerevisiae
Nature Communications (2021)
-
The polar oxy-metabolome reveals the 4-hydroxymandelate CoQ10 synthesis pathway
Nature (2021)