Abstract
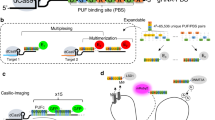
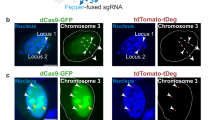
A lack of techniques to image multiple genomic loci in living cells has limited our ability to investigate chromosome dynamics. Here we describe CRISPRainbow, a system for labeling DNA in living cells based on nuclease-dead (d) Cas9 combined with engineered single guide RNA (sgRNA) scaffolds that bind sets of fluorescent proteins. We demonstrate simultaneous imaging of up to six chromosomal loci in individual live cells and document large differences in the dynamic properties of different chromosomal loci.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Hsu, P.D., Lander, E.S. & Zhang, F. Cell 157, 1262–1278 (2014).
Chen, B. et al. Cell 155, 1479–1491 (2013).
Anton, T., Bultmann, S., Leonhardt, H. & Markaki, Y. Nucleus 5, 163–172 (2014).
Ma, H. et al. Proc. Natl. Acad. Sci. USA 112, 3002–3007 (2015).
Zalatan, J.G. et al. Cell 160, 339–350 (2015).
Konermann, S. et al. Nature 517, 583–588 (2015).
Shechner, D.M., Hacisuleyman, E., Younger, S.T. & Rinn, J.L. Nat. Methods 12, 664–670 (2015).
Daigle, N. & Ellenberg, J. Nat. Methods 4, 633–636 (2007).
Austin, R.J., Xia, T., Ren, J., Takahashi, T.T. & Roberts, R.W. J. Am. Chem. Soc. 124, 10966–10967 (2002).
Jegou, T. et al. Mol. Biol. Cell 20, 2070–2082 (2009).
Smith, C.S. et al. J. Cell Biol. 209, 609–619 (2015).
Ma, H., Reyes-Gutierrez, P. & Pederson, T. Proc. Natl. Acad. Sci. USA 110, 21048–21053 (2013).
Esvelt, K.M. et al. Nat. Methods 10, 1116–1121 (2013).
Chen, B. et al. Nucleic Acids Res. http://dx.doi.org/10.1093/nar/gkv1533 (2016).
Dean, K.M. & Palmer, A.E. Nat. Chem. Biol. 10, 512–523 (2014).
Jiang, F., Zhou, K., Ma, L., Gressel, S. & Doudna, J.A. Science 348, 1477–1481 (2015).
Fu, Y., Sander, J.D., Reyon, D., Cascio, V.M. & Joung, J.K. Nat. Biotechnol. 32, 279–284 (2014).
Dahlman, J.E. et al. Nat. Biotechnol. 33, 1159–1161 (2015).
Kiani, S. et al. Nat. Methods 12, 1051–1054 (2015).
Acknowledgements
We thank C. Smith in D. Grunwald laboratory for microscopy suggestions and S.A. Wolfe, M. Fatih Bolukbasi and A. Gupta of this institution for valuable discussion. This work was supported by the Vitold Arnett Fund to T.P. and NIH grants R01 GM102515 to S.Z. and U01 EB021238 to D.G.
Author information
Authors and Affiliations
Contributions
H.M. and T.P. conceived the study and designed the experiments. H.M. and L.-C.T. performed experiments, added new innovations during the study and interpreted data. A.N. and S.Z. conducted bioinformatics mining of all the repeats in the human genome and identified the chromosome-specific repeats used as targets in this study. L.-C.T. and M.H. conducted image processing. H.M., D.G. and T.P. wrote the manuscript with the input of all the other authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Optimizing sgRNA scaffold for DNA labeling.
A) A U-A pair was mutated to G-C (shown in red at “+5”) in the sgRNA scaffold sequence. The telomere target sequence and PAM sequence are shown in green and brown respectively. The guide sequence shown in pink and the sgRNA scaffold is shown in yellow. GFP was fused to the C-terminus of Sp dCas9. B) Typical images for telomere labeling in U2OS cells by using sgRNAwt-Telo (middle left) or sgRNAplus (G-C)-Telo (right) in presence of Sp dCas9-GFP, and these two images were scaled to the same minimal and maximal levels.. Scale bar: 5 μm. Data in all panels are representative of experiments performed at least three times.
Supplementary Figure 2 Sequences and structures of CRISPRainbow sgRNAs.
RNA aptamers inserted into the telomere-targeting sgRNA were as follows: A) 2XMS2; B) 2XPP7; C) 2XboxB; D) MS2-PP7; E) PP7-boxB; F) boxB-MS2; G) boxB-MS2-PP7. The color scheme is blue for MS2, green for PP7, red for boxB, pink for guide sequences, yellow for the sgRNA scaffold, light blue for target sequence, and orange for the PAM.
Supplementary Figure 3 Optimized N22 for boxB binding and DNA labeling.
A) Two mutations were introduced into N22ori, producing N22plus. B) sgRNA-2XboxB-Telo was designed to target to telomeres, with boxB recognized by either N22ori or N22plus. C) N22ori and N22plus fused to 3XGFP were compared for the labeling of telomeres in presence of sgRNA-2XboxB-Telo and dCas9-mCherry and each column of images was scaled to the same minimal and maximal levels. Scale bar: 5 μm. Data in all panels are representative of experiments performed at least three times.
Supplementary Figure 4 Overlap of secondary colors in 3D projection.
Telomeres were labeled using sgRNAs with dual color stem loops, yellow (A), magenta (B) and cyan (C). 3D images were taken stepwise along the z axis. The step size was in 0.2 um increments. The images were registered by using Matlab software and projected using maximum projection function in Fiji software (see Methods for details). The quantification of each combinatorial color is from 6-10 cells with a total number (n) of foci analyzed: n = 208 for cyan, n = 201 for yellow and n = 195 for magenta). Scale bar: 5 μm. Data in all panels are representative of experiments performed at least three times.
Supplementary Figure 5 Dual or tricolor DNA labeling by primary colors.
Telomere and C9 were labeled by the combination of sgRNA-2XMS2-Telo / MCP-BFP and sgRNA-2XPP7-C9 / PCP-GFP (A), sgRNA-2XPP7-Telo / PCP-GFP and sgRNA-2XboxB-C9 / N22-RFP (B) or sgRNA-2XboxB-Telo / N22-RFP and sgRNA-2XMS-C9 / MCP-BFP (C) in the presence of dCas9. Scale bar: 5 μm. D) DNA labeling by three primary colors. A repeated sequence unique to human chromosome 9, another unique to chromosome 3 (see text and Supplemental Table 2) and telomeres were labeled by co-expression of dCas9, the three indicated sgRNAs, and the three cognate fluorescent proteins. Scale Bar: 5 μm. Data in all panels are representative of experiments performed at least three times.
Supplementary Figure 6 Short sgRNA for DNA labeling.
A) The guide sequences termed C3-20 mer, C3-11 mer, C13-20 mer and C13-11 mer were cloned into pLH-sgRNA for DNA labeling. B) Typical images for C3 labeling by either sgRNA with C3-20 mer (middle left) or C3-11 mer (right) in U2OS cells in presence of Sp dCas9-GFP and these two images were scaled to the same minimal and maximal levels. Scale bar: 5 μm. C) Typical images for C13 labeling by either sgRNA with C13-20 mer (middle left) or C13-11 mer (right) in U2OS cells in presence of Sp dCas9-GFP and these two images were scaled to the same minimal and maximal levels. All the foci were indicated by white arrows. Scale bar: 5 μm. D) Four-color DNA labeling. C9, C13 and telomere used primary colors, while C3 locus labeled in this case using a sgRNA carrying both the PP7 and boxB elements. Shown is a cell following co-expression of dCas9, the three sgRNAs and the cognate fluorescent proteins. As can be seen in the merge image, three yellow loci are evident (yellow arrows), representing the C3 locus Scale bar: 3 μm. Data in all panels are representative of experiments performed at least three times.
Supplementary Figure 7 Chromosome-specific targets for primary colors sand secondary colors.
Chromosome-specific targets were labeled by three primary colors (A) and three secondary colors (B) by co-expression of dCas9, the three indicated sgRNAs, and the three cognate fluorescent proteins. Scale bar: 3 μm. Data are representative of experiments performed at least three times.
Supplementary Figure 8 Trajectory overlay of chromosome movement in time-lapse.
Overlay of fluorescent foci with bright-field image (A) is shown. A total of six foci including four different labeling sites: chromosome 3 (C3-1, C3-2, C3-3, yellow), chromosome 9 (C9, blue), chromosome 13 (C13, green) and telomeres (Telo, red) were chosen. Trajectory overlay of three chromosome 3 foci and all foci are shown in B and C respectively. Images were continuously taken at the rate of 20 frames per second (fps) over 10 seconds. The bright-field image was registered and contrasted as described in Methods except an additional background subtraction process, viz. the flatten background function in Metamorph was applied for better visibility. All trajectories are mathematically shifted to start from the origin (0, 0) for easy comparison and readability. Scale bar: 5 μm. Data are representative of experiments performed at least three times.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 and Supplementary Tables 1 and 2 (PDF 2549 kb)
41587_2016_BFnbt3526_MOESM20_ESM.mov
Time-lapse of four DNA loci movements: Unique sites in chromosome 3, 9 and 13 as well as telomeres were labeled simultaneously using the CRISPRainbow colors blue, green, red and yellow respectively. The movements of these loci were recorded at 50 ms per frame for 10 seconds (200 total frames). (MOV 3163 kb)
Rights and permissions
About this article
Cite this article
Ma, H., Tu, LC., Naseri, A. et al. Multiplexed labeling of genomic loci with dCas9 and engineered sgRNAs using CRISPRainbow. Nat Biotechnol 34, 528–530 (2016). https://doi.org/10.1038/nbt.3526
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.3526
This article is cited by
-
Spatiotemporal control of RNA metabolism and CRISPR–Cas functions using engineered photoswitchable RNA-binding proteins
Nature Protocols (2024)
-
3D genomics and its applications in precision medicine
Cellular & Molecular Biology Letters (2023)
-
Live-cell imaging of chromatin contacts opens a new window into chromatin dynamics
Epigenetics & Chromatin (2023)
-
Recent advances and applications of CRISPR-Cas9 in cancer immunotherapy
Molecular Cancer (2023)
-
CRISPR imaging reveals chromatin fluctuation at the centromere region related to cellular senescence
Scientific Reports (2023)