Abstract
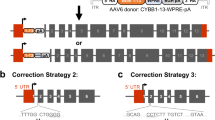
Gene therapy with genetically modified human CD34+ hematopoietic stem and progenitor cells (HSPCs) may be safer using targeted integration (TI) of transgenes into a genomic 'safe harbor' site rather than random viral integration. We demonstrate that temporally optimized delivery of zinc finger nuclease mRNA via electroporation and adeno-associated virus (AAV) 6 delivery of donor constructs in human HSPCs approaches clinically relevant levels of TI into the AAVS1 safe harbor locus. Up to 58% Venus+ HSPCs with 6–16% human cell marking were observed following engraftment into mice. In HSPCs from patients with X-linked chronic granulomatous disease (X-CGD), caused by mutations in the gp91phox subunit of the NADPH oxidase, TI of a gp91phox transgene into AAVS1 resulted in ∼15% gp91phox expression and increased NADPH oxidase activity in ex vivo–derived neutrophils. In mice transplanted with corrected HSPCs, 4–11% of human cells in the bone marrow expressed gp91phox. This method for TI into AAVS1 may be broadly applicable to correction of other monogenic diseases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Biffi, A. et al. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science 341, 1233158 (2013).
Hacein-Bey-Abina, S. et al. Efficacy of gene therapy for X-linked severe combined immunodeficiency. N. Engl. J. Med. 363, 355–364 (2010).
Cavazzana-Calvo, M. et al. Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science 288, 669–672 (2000).
Hacein-Bey-Abina, S. et al. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N. Engl. J. Med. 348, 255–256 (2003).
Stein, S. et al. Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease. Nat. Med. 16, 198–204 (2010).
Carroll, D. Genome engineering with targetable nucleases. Annu. Rev. Biochem. 83, 409–439 (2014).
Tebas, P. et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N. Engl. J. Med. 370, 901–910 (2014).
DeKelver, R.C. et al. Functional genomics, proteomics, and regulatory DNA analysis in isogenic settings using zinc finger nuclease-driven transgenesis into a safe harbor locus in the human genome. Genome Res. 20, 1133–1142 (2010).
Genovese, P. et al. Targeted genome editing in human repopulating haematopoietic stem cells. Nature 510, 235–240 (2014).
Carroll, D. & Beumer, K.J. Genome engineering with TALENs and ZFNs: repair pathways and donor design. Methods 69, 137–141 (2014).
Urnov, F.D. et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 435, 646–651 (2005).
Moehle, E.A. et al. Targeted gene addition into a specified location in the human genome using designed zinc finger nucleases. Proc. Natl. Acad. Sci. USA 104, 3055–3060 (2007).
Vierstra, J. et al. Functional footprinting of regulatory DNA. Nat. Methods 12, 927–930 (2015).
Song, L. et al. Optimizing the transduction efficiency of capsid-modified AAV6 serotype vectors in primary human hematopoietic stem cells in vitro and in a xenograft mouse model in vivo. Cytotherapy 15, 986–998 (2013).
Song, L. et al. High-efficiency transduction of primary human hematopoietic stem cells and erythroid lineage-restricted expression by optimized AAV6 serotype vectors in vitro and in a murine xenograft model in vivo. PLoS One 8, e58757 (2013).
Orlando, S.J. et al. Zinc-finger nuclease-driven targeted integration into mammalian genomes using donors with limited chromosomal homology. Nucleic Acids Res. 38, e152 (2010).
Kuhns, D.B. et al. Residual NADPH oxidase and survival in chronic granulomatous disease. N. Engl. J. Med. 363, 2600–2610 (2010).
Marciano, B.E. et al. Common severe infections in chronic granulomatous disease. Clin. Infect. Dis. 60, 1176–1183 (2015).
Challita, P.M. et al. Multiple modifications in cis elements of the long terminal repeat of retroviral vectors lead to increased expression and decreased DNA methylation in embryonic carcinoma cells. J. Virol. 69, 748–755 (1995).
Astrakhan, A. et al. Ubiquitous high-level gene expression in hematopoietic lineages provides effective lentiviral gene therapy of murine Wiskott-Aldrich syndrome. Blood 119, 4395–4407 (2012).
Wang, J. et al. Homology-driven genome editing in hematopoietic stem and progenitor cells using ZFN mRNA and AAV6 donors. Nat. Biotechnol. 33, 1256–1263 (2015).
Langmead, B. & Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Gabriel, R. et al. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat. Biotechnol. 29, 816–823 (2011).
Brenner, S. et al. Concentrated RD114-pseudotyped MFGS-gp91phox vector achieves high levels of functional correction of the chronic granulomatous disease oxidase defect in NOD/SCID/β2-microglobulin−/− repopulating mobilized human peripheral blood CD34+ cells. Blood 102, 2789–2797 (2003).
Acknowledgements
These studies were supported in part by project 1 ZIA AI000644 of the intramural program of National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health. Human CD34+ HSPCs were obtained under NIAID IRB approved Protocol 94-I-0073 after written informed consent. Murine animal studies were performed under National Institutes of Allergy and Infectious Diseases (NIAID) Animal Care and Use Committee approved protocol Laboratory of Host Defenses (LHD) 3E.
Author information
Authors and Affiliations
Contributions
This research was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health under intramural project numbers Z01-AI-00644 and Z01-AI-00988. S.S.D.R., A.R., P.-Q.L., L.L. and X.W. performed most of the experiments, N.T., U.C., J.L., S.K., C.R., H.N., L.S., C.S., A.H.S., A.C., J.R.P., D.E.P. and D.A.S. developed reagent, ran assays and analyzed samples, M.V.P. and P.D.G. provided support, S.S.D.R., K.A.Z., A.R., F.D.U. and H.L.M. designed experiments and wrote manuscript.
Corresponding author
Ethics declarations
Competing interests
The following authors are full-time employees of Sangamo BioSciences and might own Sangamo stock or derivatives: A.R., P.-Q.L., C.R., A.H.S., A.C., J.R.P., D.E.P., D.A.S., P.D.G. and F.D.U. L.L. and M.V.P. are full-time employees of MaxCyte Systems.
Integrated supplementary information
Supplementary Figure 1 AAVS1 Venus TI with varying AAVS1 ZFN mRNA amount and AAV6 Venus MOI
To extend conditions for transfection, experiments as described in Fig 2 were repeated using lower concentrations of AAVS1 ZFN mRNA as indicated, immediately followed by exposure to indicated AAV6 Venus donor MOIs. This figure shows cell viability (top), percent of live cells expressing Venus marker (middle) and relative viable cell numbers at each day of culture after treatment in each sample following treatment (bottom). Values were determined for each analysis at culture day 3, 4, 6 and 9 (corresponding to 1, 2, 4 and 7 days post treatment). The data demonstrated the significant impact of titrating amount of ZFN mRNA to achieve optimal outcome, which for the ZFN mRNAs used in our study, was achieved with 25gμg/mL.
Supplementary Figure 2 Optimization of culture days before treatment.
Following thaw (day 0), healthy volunteer CD34+ HSC were cultured for a period of 1, 2 or 3 days before treatment with electroporation delivery of AAVS1 ZFN mRNA followed by AAV6 Venus donor addition. This figure shows cell viability (top), and percent of live cells expressing Venus marker % Venus+ cells in the gated live cells at 2, 3, 4, and 6 days following treatment (blue, red, green and black bars respectively).
Supplementary Figure 3 Schema for molecular analysis for TI.
a. Out/Out PCR utilizes primers located outside the donor homology arm (green) with HDR-F4 and HDR-R5; Out/In PCR with one primer situated outside the left homology arm and the second primer within the construct (HDR-F4 and 2A-R (or gp91-R)), or within the construct to the right homology arm, ie the In/Out PCR (primers 2A-F (for gp91-F) and HDR-R5). The regular MiSeq is performed with primers Mi-F and Mi-R; 5’-2A-TI Miseq with primers Mi-F, 2A-R, and Mi-R; and 3’MiSeq with primers Mi-F, polyA-F, and Mi-R.
b. Schema showing the primers for extended sequencing of the AAVS1 TI junction by PacBio single molecule sequencing. PCR primer pairs with one inside the vector specific region and the other outside the vector in the non-overlapping genomic region of the AAVS1 locus results in amplification of the junctions of the integrated vector.
Supplementary Figure 4 Representative sequences of AAV6 TI junction at the AAVS1 locus.
Representative sequences showing the junctions of AAV6 Venus integration at the AAVS1 locus by HDR (i, ii) or NHEJ (iii). A representative sequence for the left arm (L) junction at the AAVS1 locus following TI by HDR shows AAVS1 sequences 5’ beyond the AAV6 vector-L arm AAVS1 junction-AAV6 vector specific sequence (i). A representative sequence of the R arm junction is shown as AAV6 vector specific sequence-R arm AAVS1 junction-AAVS1 sequence 3’ beyond the vector (ii). A representative sequence of a NHEJ-mediated junction is shown as AAVS1 sequence 5’ shared AAVS1 and AAV6 vector-AAVS1 sequence shared by L arm of AAV6 vector-ITR-L arm of AAVS1 seq shared with AAV6 vector-AAV6 vector specific sequence (iii). Since the ITR lies outside the homology region, it is not incorporated during HDR so that the junction sequence does not contain the ITR. However, during NHEJ, the ITR sequence is retained in the junction. The expected amplicon sizes range from 1.1 kb to 2.5 kb depending on whether the ITR is retained. Both AAVS1L junction and AAVS1R junction were amplified, and PacBio long single DNA molecule sequencing identified 1082 integration events without ITR sequence and 3 integration events retaining ITR sequence. The results suggest that almost all AAV integration at the AAVS1 locus we observed occurred through HDR between AAV6 vector bearing-AAVS1 sequence flanking the transgene and endogenous AAVS1 sequence.
Supplementary Figure 5 Mouse transplant study analysis at 8 weeks.
Insert figure caption here by deleting or overwriting this text; captions may run to a second page if necessary. To ensure accurate appearance in the published version, please use the Symbol font for all symbols and Greek letters.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5 and Supplementary Table 1 (PDF 1886 kb)
Rights and permissions
About this article
Cite this article
De Ravin, S., Reik, A., Liu, PQ. et al. Targeted gene addition in human CD34+ hematopoietic cells for correction of X-linked chronic granulomatous disease. Nat Biotechnol 34, 424–429 (2016). https://doi.org/10.1038/nbt.3513
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.3513
This article is cited by
-
Gene Editing in Human Haematopoietic Stem Cells for the Treatment of Primary Immunodeficiencies
Molecular Diagnosis & Therapy (2023)
-
Gene-Modified Stem Cells for Spinal Cord Injury: a Promising Better Alternative Therapy
Stem Cell Reviews and Reports (2022)
-
Correction to: Treating primary immunodeficiencies with defects in NK cells: from stem cell therapy to gene editing
Stem Cell Research & Therapy (2021)
-
Homology-directed gene-editing approaches for hematopoietic stem and progenitor cell gene therapy
Stem Cell Research & Therapy (2021)
-
Generation of somatic mitochondrial DNA-replaced cells for mitochondrial dysfunction treatment
Scientific Reports (2021)