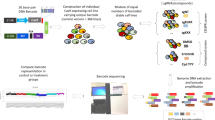
Abstract
Hundreds of genetically characterized cell lines are available for the discovery of genotype-specific cancer vulnerabilities. However, screening large numbers of compounds against large numbers of cell lines is currently impractical, and such experiments are often difficult to control1,2,3,4. Here we report a method called PRISM that allows pooled screening of mixtures of cancer cell lines by labeling each cell line with 24-nucleotide barcodes. PRISM revealed the expected patterns of cell killing seen in conventional (unpooled) assays. In a screen of 102 cell lines across 8,400 compounds, PRISM led to the identification of BRD-7880 as a potent and highly specific inhibitor of aurora kinases B and C. Cell line pools also efficiently formed tumors as xenografts, and PRISM recapitulated the expected pattern of erlotinib sensitivity in vivo.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
McDermott, U. et al. Identification of genotype-correlated sensitivity to selective kinase inhibitors by using high-throughput tumor cell line profiling. Proc. Natl. Acad. Sci. USA 104, 19936–19941 (2007).
Sos, M.L. et al. Predicting drug susceptibility of non-small cell lung cancers based on genetic lesions. J. Clin. Invest. 119, 1727–1740 (2009).
Barretina, J. et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 (2012).
Garnett, M.J. et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 483, 570–575 (2012).
Sharma, S.V., Haber, D.A. & Settleman, J. Cell line-based platforms to evaluate the therapeutic efficacy of candidate anticancer agents. Nat. Rev. Cancer 10, 241–253 (2010).
Abaan, O.D. et al. The exomes of the NCI-60 panel: a genomic resource for cancer biology and systems pharmacology. Cancer Res. 73, 4372–4382 (2013).
Basu, A. et al. An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell 154, 1151–1161 (2013).
Turner, N.C. & Reis-Filho, J.S. Genetic heterogeneity and cancer drug resistance. Lancet Oncol. 13, e178–e185 (2012).
Peck, D. et al. A method for high-throughput gene expression signature analysis. Genome Biol. 7, R61 (2006).
Du, J. et al. Bead-based profiling of tyrosine kinase phosphorylation identifies SRC as a potential target for glioblastoma therapy. Nat. Biotechnol. 27, 77–83 (2009).
Muellner, M.K. et al. A chemical-genetic screen reveals a mechanism of resistance to PI3K inhibitors in cancer. Nat. Chem. Biol. 7, 787–793 (2011).
Koivunen, J.P. et al. EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin. Cancer Res. 14, 4275–4283 (2008).
Wilhelm, S.M. et al. Preclinical overview of sorafenib, a multikinase inhibitor that targets both Raf and VEGF and PDGF receptor tyrosine kinase signaling. Mol. Cancer Ther. 7, 3129–3140 (2008).
Flaherty, K.T. et al. METRIC Study Group. Improved survival with MEK inhibition in BRAF-mutated melanoma. N. Engl. J. Med. 367, 107–114 (2012).
Comer, E. et al. Fragment-based domain shuffling approach for the synthesis of pyran-based macrocycles. Proc. Natl. Acad. Sci. USA 108, 6751–6756 (2011).
Lowe, J.T. et al. Synthesis and profiling of a diverse collection of azetidine-based scaffolds for the development of CNS-focused lead-like libraries. J. Org. Chem. 77, 7187–7211 (2012).
Marcaurelle, L.A. et al. An aldol-based build/couple/pair strategy for the synthesis of medium- and large-sized rings: discovery of macrocyclic histone deacetylase inhibitors. J. Am. Chem. Soc. 132, 16962–16976 (2010).
Schreiber, S.L. et al. Cancer Target Discovery and Development Network. Towards patient-based cancer therapeutics. Nat. Biotechnol. 28, 904–906 (2010).
Zhang, X.D. Illustration of SSMD, z score, SSMD*, z* score, and t statistic for hit selection in RNAi high-throughput screens. J. Biomol. Screen. 16, 775–785 (2011).
Andrews, P.D., Knatko, E., Moore, W.J. & Swedlow, J.R. Mitotic mechanics: the auroras come into view. Curr. Opin. Cell Biol. 15, 672–683 (2003).
Carmena, M. & Earnshaw, W.C. The cellular geography of aurora kinases. Nat. Rev. Mol. Cell Biol. 4, 842–854 (2003).
Ditchfield, C. et al. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 161, 267–280 (2003).
Davis, M.I. et al. Comprehensive analysis of kinase inhibitor selectivity. Nat. Biotechnol. 29, 1046–1051 (2011).
Karaman, M.W. et al. A quantitative analysis of kinase inhibitor selectivity. Nat. Biotechnol. 26, 127–132 (2008).
Korn, J.M. et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat. Genet. 40, 1253–1260 (2008).
Acknowledgements
We thank S. Kim, G. Bonamy, J. (J.) Che, J. Thibault, T. Huynh, I. Engels and A. Shipway at Novartis for sharing data before publication; A. Christie and T. Davis for technical assistance in animal studies; C. Hartland, S. Donovan, E. Rubin and E. Winchester for technical assistance in compound assays; J. Bittker, J. McGrath and G. Wendel for assistance in compound management; S. Le Quement and J. Duvall for assistance in compound synthesis; J. Gale for technical assistance in enzyme kinetic assays; S. Howell for assistance in curation of computational data sets; A. Koehler, S. Dandapani, B. Muñoz, C. Scherer, D. Gray, D. Bachovchin, S. Santaguida and J. Elkins for expert scientific guidance; J. Barretina, N. Stransky, S. Nijman, B. Julian, W. Read-Button, J. Davis and D. Peck for technical advice; and members of the Golub laboratory for critical review of the manuscript. This work was supported in part by the US National Institutes of Health (NIH) Genomics Based Drug Discovery consortium grants RL1-CA133834, RL1-GM084437 and UL1DE019585 (administratively linked to NIH grant RL1-HG004671), US National Cancer Institute Integrative Cancer Biology Program grant U54CA112962, the Howard Hughes Medical Institute, the Claudia Adams Barr Program in Cancer Research Innovative Basic Science Research Program Grant, the American Society of Clinical Oncology Conquer Cancer Foundation Young Investigator Award and the Prostate Cancer Foundation.
Author information
Authors and Affiliations
Contributions
C.Y. and T.R.G. designed the PRISM method and wrote the manuscript. C.Y., G.M.Y. and A.M.M. performed the experiments in the study. L.A.G. provided cell lines and drug response validation data from the Cancer Cell Line Encyclopedia Project. B.A.W. performed cell line genotype verification analyses. K.N.R. and P.T. contributed to statistical analyses of PRISM validation data. J.Z.G. and C.Y. created data processing and data visualization tools. M.A.P., W.W., A.C., M.A.M., B.R.T., G.M.Y., A.M.M. and C.Y. performed the large-scale PRISM screen. A.T. and C.Y. performed genomic correlation analyses in the large-scale PRISM screen. A.F.S. and S.L.S. contributed to compound creation and curation and design of experiments with BRD-7880. Y.-L.Z. performed kinetic kinase inhibition experiments with BRD-7880. A.L.K., C.Y. and T.R.G. contributed to design and execution of in vivo PRISM experiments. B.W. contributed to data visualization tools and to manuscript figures. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
C.Y. and T.R.G. are inventors in patent application PCT/US2013/031312.
Integrated supplementary information
Supplementary Figure 1 Response to erlotinib measured in pools and in individual cell lines.
Relative cell number after 5-day exposure of cell lines to erlotinib was determined in mixture (PRISM) or with randomly selected individual cell lines harboring wild-type (wt) or mutant (mut) EGFR using CellTiter Glo (ATP). At each concentration the mean relative cell viability (DMSO-treated cells = 100%) is plotted with error bars reflecting standard error of the mean. C, control (DMSO) treatment (no erlotinib). Right, IC50 determinations for each method.
Supplementary Figure 2 Selective drug action in cell lines detected by PRISM.
a. Hierarchical clustering of 43 anticancer and control compounds based on AUC measurements of 100 cell lines performed with PRISM.
b. Comparison of genotype-predicted responses of cell lines to targeted anticancer drugs, as determined by three cell viability measures.
Oncogene mutations in the BRAF genes were determined previously for 100 cell lines3 and used to stratify responses to specific compounds shown. Boxplots show medians, 25th, and 75th percentiles, with bars showing standard error of the mean. PRISM, Nuclei, and ATP demonstrated significant (two-tailed t-test, asterisk denotes p < 0.05) reductions in cell viability with the BRAF inhibitor PLX4720 in BRAF V600E mutant lines compared to BRAF wild-type lines; a similar reduction was seen only with PRISM with the RAF inhibitor RAF-265. The RAF inhibitor sorafenib did not demonstrate significant reductions in BRAF-mutant vs. -wild-type lines. The MEK inhibitor AZD6244 demonstrated significant reduction in BRAF V600E mutant lines compared to BRAF wild-type lines with ATP and a trend towards this reduction in both PRISM (p = 0.054) and Nuclei (p = 0.099).
Supplementary Figure 3 Outlier analysis of discrepancies between PRISM and other cell viability measurements.
The log2 of the ratio of the AUC measured by PRISM to the AUC measured by either Nuclei (a) or ATP (b) was used to systematically determine whether specific cell lines (shown in columns) or compounds (shown in rows) were enriched for wider discrepancies between PRISM and other cell viability measurements. Discordances in the heat map are shown in blue (where AUCPRISM is less than AUCNuclei) or red (where the converse is true).Yellow shading denotes assays which were not performed in respective Nuclei or ATP analyses.
Supplementary Figure 4 Relative tumor cell line growth in mixture in vivo using PRISM.
A tumor from a randomly selected animal treated with vehicle was cut into four sections to examine for variance in cell line composition across the tumor. Abundance of each cell line within a particular section, as determined by PRISM, is plotted as a relative percentage of the tumor section.
Supplementary Figure 5 Relative growth of barcoded cell lines in vitro over time.
Twenty-four barcoded lung adenocarcinoma cell lines were mixed in equal numbers and grown in culture with weekly passage over 98 days. Genomic DNA harvested from a sample of each week’s mixture was analyzed with PRISM to determine the relative proportions of each cell line in the mixture.
Supplementary Figure 6 Distribution of Luminex signals in vehicle-treated and compound-treated barcoded cell lines in PRISM.
a. Mean Luminex signals minus background of 102 barcoded cell lines treated with DMSO vehicle, with error bars reflecting the standard error of the mean. Numbers in brackets denote the barcode identifier.
b. Distribution of scaled Luminex signals of individual barcoded cell lines across 400 compounds tested at dose. Box plots show mean and 25th and 75th percentiles; whiskers show highest and lowest values. Full dataset is in Supplementary Table 3.
c. Cell equivalents of each cell line were calculated by dividing the mean Luminex signals minus background in DMSO-treated control samples at the end of the assay by the corresponding signal from a control lysate containing equal numbers of each cell line. The cell equivalent for each cell line was compared as a ratio to the median cell equivalent to show the range of cell numbers across cell lines at the end of the assay.
d. Correlation of Luminex signal of DMSO-treated control for each cell line with doubling time.
Supplementary Figure 7 Strictly standardized mean differences in 102 barcoded cell lines in PRISM.
The strictly standardized mean difference (β) robust for outliers20 was estimated using
Supplementary Figure 8 Hierarchical clustering of bioactive compounds by activity against 102 barcoded cell lines.
Spearman-based hierarchical clustering was used to determine compounds with the most similar patterns of activity against 102 barcoded cell lines in PRISM. Four highly related clusters of activity each demonstrate compounds with similar modes of action (from left): corticosteroids, HMG-CoA reductase inhibitors, EGFR inhibitors, mTOR inhibitors, and EGFR inhibitors. The full heatmap with 400 compounds (with associated data in Supplementary Table 3) is included as digital Supplementary Figure 8b.
Provided separately in digital file format as Suppl_Figure_8b_GOLUB.pdf
Supplementary Figure 9 Hierarchical clustering of barcoded cell line responses to treatment with 400 bioactive compounds.
Pearson-based hierarchical clustering was used to determine cell lines with the most similar responses to bioactive compounds using PRISM. Pairs of cell lines with matching genetic identities by SNP fingerprinting but with distinct barcodes are denoted with matching colored arrows. 5 of 6 pairs of cell lines demonstrated marked similarity by clustering. Of note the cell line PC-14 has been reported to be identical to PC-9 (http://www.brc.riken.jp/lab/cell/english/rcb0446_announce.shtml). Full heatmap is in digital Supplementary Figure 8b.
Supplementary Figure 10 BRD-7880 inhibits activities of aurora kinase B.
BRD-7880 and other aurora kinase inhibitors increase DNA content of HCT-116 cells. HCT-116 cells were treated with 10 µM of DMSO, barasertib, GSK-1070916, MLN8054, BRD-7880, or tozasertib. 24 hours or 48 hours following treatment, cells were stained with propidium iodide and DNA content per cell was assessed by flow cytometry. Vertical dashed line represents diploid DNA content in DMSO-treated cells.
BRD-7880 and other aurora kinase inhibitors decrease phosphorylation of serine 10 on histone H3, a marker of AURKB kinase activity. HCT-116 cells were treated with 10 µM of DMSO, barasertib, GSK1070916, MLN8054, BRD-7880, or tozasertib. Cells lysates were probed on Western blot using antibodies to histone H3, phosphoserine10-histone H3, AURKB, or beta-actin and detected using a LI-COR Odyssey analyzer.
Supplementary Figure 11 Profiles of aurora kinase inhibitors in in vitro aurora kinase assays.
Incorporation of radioactivity from 10 µM γ-33P-ATP was measured in in vitro kinase assays across 8 doses of various aurora kinase inhibitors in duplicate by the EMD Millipore KinaseProfiler service under published standard conditions with ATP at Km. Full-length human AURKB was assayed with 30 µM AKRRRLSSLRA (ribosomal protein S6 peptide). IC50 values were modeled using least-squares and variable slope with GraphPad Prism 6.0 software.
Supplementary Figure 12 BRD-7880, tozasertib, and barasertib are ATP-competitive inhibitors of AURKB In vitro.
Enzyme kinetic experiments were performed with recombinant AURKB with fluorescently labeled Caliper peptide substrate with various concentrations of ATP and compound (BRD-7880, tozasertib, barasertib), and fluorescent product and substrate were separated and monitored on a Perkin-Elmer Caliper LabChip EZ Reader. Km and ki values were determined from the double reciprocal Lineweaver-Burk plot by linear regression with GraFit 6 software using competitive inhibition equation modeling.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–2 and Supplementary Methods, Supplementary Table 8 and 9 (PDF 6692 kb)
Suppl_Figure_8b_GOLUB.pdf
Hierarchical clustering of bioactive compounds by activity against 102 barcoded cell lines. (PDF 830 kb)
Supplementary Table 1
PRISM AUC (Area Under the Curve) measurements of cell viability (XLS 117 kb)
Supplementary Table 2
PRISM cell viability of 8,000 Diversity-Oriented Synthesis (DOS) compounds at single dose across 102 cell lines (XLS 15752 kb)
Supplementary Table 3
PRISM cell viability across 8 or more doses of 400 tool compounds or oncology drugs with known mechanism-of-action and BRD-7880 (XLS 7440 kb)
Supplementary Table 4
SNP fingerprinting of 102 cell lines (XLS 141 kb)
Supplementary Table 5
Genomic correlates of PRISM sensitivity profiles (ZIP 169492 kb)
Supplementary Table 6
PRISM cell viability across 8 doses of 199 DOS compounds (XLS 3002 kb)
Supplementary Table 7
PRISM cell viability across 8 doses of tozasertib and BRD-7880 (XLS 132 kb)
Rights and permissions
About this article
Cite this article
Yu, C., Mannan, A., Yvone, G. et al. High-throughput identification of genotype-specific cancer vulnerabilities in mixtures of barcoded tumor cell lines. Nat Biotechnol 34, 419–423 (2016). https://doi.org/10.1038/nbt.3460
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.3460
This article is cited by
-
Inflammatory response-based prognostication and personalized therapy decisions in clear cell renal cell cancer to aid precision oncology
BMC Medical Genomics (2023)
-
COMMD3 loss drives invasive breast cancer growth by modulating copper homeostasis
Journal of Experimental & Clinical Cancer Research (2023)
-
Multi-omics analysis uncovers clinical, immunological, and pharmacogenomic implications of cuproptosis in clear cell renal cell carcinoma
European Journal of Medical Research (2023)
-
Combined in vitro/in vivo genome-wide CRISPR screens in triple negative breast cancer identify cancer stemness regulators in paclitaxel resistance
Oncogenesis (2023)
-
Rewiring cancer drivers to activate apoptosis
Nature (2023)