Abstract
The ability to computationally predict the effects of toxic compounds on humans could help address the deficiencies of current chemical safety testing. Here, we report the results from a community-based DREAM challenge to predict toxicities of environmental compounds with potential adverse health effects for human populations. We measured the cytotoxicity of 156 compounds in 884 lymphoblastoid cell lines for which genotype and transcriptional data are available as part of the Tox21 1000 Genomes Project. The challenge participants developed algorithms to predict interindividual variability of toxic response from genomic profiles and population-level cytotoxicity data from structural attributes of the compounds. 179 submitted predictions were evaluated against an experimental data set to which participants were blinded. Individual cytotoxicity predictions were better than random, with modest correlations (Pearson's r < 0.28), consistent with complex trait genomic prediction. In contrast, predictions of population-level response to different compounds were higher (r < 0.66). The results highlight the possibility of predicting health risks associated with unknown compounds, although risk estimation accuracy remains suboptimal.
Similar content being viewed by others
Main
The ability to predict a toxic response in a population could help establish safe levels of exposure to new compounds and identify individuals at increased risk for adverse health outcomes. Current risk assessment does not account for individual differences in chemical exposure response. Furthermore, standard safety testing is performed on a small fraction of existing environmental compounds1 and uses animal models that are costly, time-consuming and do not always reflect human safety profiles2. Algorithms that provide accurate in silico predictions of safety risks in humans could provide an accurate and cost-effective tool to identify potential health risks to specific populations. However, previous prediction algorithms have been limited by lack of data about population variability and difficulties in extrapolating from model organisms3,4.
The development of high-throughput in vitro toxicity studies using human-derived cell models5 and rapidly decreasing sequencing costs have enabled large, genetically distinct populations to be characterized. High-throughput in vitro systems have been successfully used to assess changes in transcriptional6,7 and phenotypic8 traits in response to compound exposure. Furthermore, genomically characterized cell lines that decrease nongenetic sources of variation9,10 have been used to identify genetic variants and transcripts associated with both in vitro and clinical responses to drug exposures11,12. These technologies enable systematic toxicity screening of a wide range of compounds in human cell lines to assess population-level responses and to examine variation in risk profiles across individuals13.
This work formed part of an open community challenge within the Dialogue on Reverse Engineering Assessment and Methods (DREAM) framework14,15. Participating researchers were asked to predict interindividual variability in cytotoxic response based on genomic and transcriptional profiles (subchallenge 1) and to predict population-level parameters of cytotoxicity across chemicals based on structural attributes of compounds (subchallenge 2). Cellular toxicity was assessed for 156 compounds across lymphoblastoid cell lines derived from 884 individuals5 from nine distinct geographical subpopulations across Europe, Africa, Asia and the Americas (Fig. 1)16. Genetic17 and transcriptional data18 from these cell lines were available as part of the 1000 Genomes Project. The data set has twice the number of cell lines and three times the number of compounds compared with the previous largest study19. We evaluated the submitted state-of-the-art modeling approaches to benchmark current best practices in predictive modeling. Furthermore, the challenge identified algorithms that were able to predict, with better than random accuracy, individual and population-level response to different compounds using only genomic data. Although these results represent an improvement over previous attempts to predict cytotoxicity response, substantial improvements in prediction accuracy remain critical.
The cytotoxicity data used in the challenge consist of the EC10 data generated for 884 lymphoblastoid cell line in response to 156 common environmental compounds. Participants were provided with a training set of cytotoxicity data for 620 cell lines and 106 compounds along with genotype data for all cell lines, RNA-seq data for 337 cell lines and chemical attributes for all compounds. The challenge was divided into two independent subchallenges: in subchallenge 1, participants were asked to predict EC10 values for a separate test set of 264 cell lines in response to the 106 compounds (only 91 toxic compounds were used for final scoring); in subchallenge 2, they were asked to predict population parameters (in terms of median EC10 values and 5th (q05) to 95th (q95) interquantile distance) for a separate test set of 50 compounds.
Results
Challenge participation
213 people from more than 30 countries registered to participate in the NIEHS-NCATS-UNC (National Institute of Enivronmental Health Sciences–National Center for Advancing Translational Sciences–University North Carolina) DREAM Toxicogenetics challenge. Participants were provided with a subset of the data to train models over a 3-month period, and models were evaluated on a second subset of test data to which the participants were blinded (Fig. 1). The training data included (i) a measure of cytotoxic susceptibility per cell line (EC10, the dose for which a 10% decrease in viability occurred) for 106 compounds tested across 487 cell lines; (ii) genotypes for all 884 cell lines; (iii) RNA-seq-based quantification of gene transcripts for 337 cell lines, and (iv) structural attributes of all 156 compounds. 34 research teams submitted a total of 99 predictions of interindividual variation in response to subchallenge 1, and 23 research teams submitted a total of 80 predictions of population-level toxicity parameters in response to subchallenge 2. The challenge offered the unique opportunity to compare performance across a wide variety of state-of-the-art methods (Supplementary Table 1 and Supplementary Predictive Models) for the prediction of cytotoxic response to environmental compounds.
Subchallenge 1: prediction of interindividual variation
Models were evaluated based on their ability to predict EC10 values in a blind test set comprising EC10 values measured in 264 cell lines that were not included in the training set. Prediction accuracy was scored using two metrics: Pearson correlation (r), which evaluates the linear dependence between predicted and measured EC10 values, and a rank-based metric, the probabilistic C-index (pCi)15, which takes into account the probabilistic nature of the gold standard due to technical sources of noise in the associated measures by evaluating the concordance of cell line cytotoxicity ranks. Scoring analyses were limited to 91 compounds, excluding 15 compounds for which no effect on cytotoxicity was observed across the entire population, in order to avoid the introduction of noise in the ranking (Supplementary Fig. 1). For each metric, overall team ranks were calculated by ranking teams separately for each compound and then averaging across compounds.
We first assessed whether predictions were significantly better than random by comparing the average r (Fig. 2a) and the average pCi (Fig. 2b), computed across compounds for each submission with the corresponding null model of randomly empirically generated EC10 values. Out of the 99 submissions, the null hypothesis of randomly generated predictions could be rejected (false discovery rate (FDR) < 0.05, which automatically corrects for multiple hypotheses) for 46 submissions using r, 47 submissions using pCi and for 42 submissions using both metrics. The average values over all compounds for r and pCi were quite modest (maximum value 0.07 and 0.51, respectively) suggesting that cytotoxic response to chemical exposure is not, in general, well predicted based on single-nucleotide polymorphism (SNP) data. Although average predictive ability was low, performance was not uniform across compounds. Variability in predictive performance across compounds ranged from −0.21 to 0.28 for r values and from 0.45 to 0.56 for pCi (Fig. 3a and Supplementary Fig. 2). We tested whether cytotoxicity of each compound could be predicted better than chance. For each compound, predicted EC10 values for all teams were compared with the null random model. This analysis verified that predictions are significantly better than random for most of the compounds (55 out of the 91 compounds; Wilcoxon rank sum test, P < 0.05), even if performances for some compounds are very poor. The ranking of best performing teams was shown to be robust with respect to the compounds used for scoring (Supplementary Fig. 3).
(a–d) Submissions are compared with the null hypothesis for subchallenge 1 (a,b) and subchallenge 2 (c,d). For each metric used for scoring (Pearson correlation (a) and pCi (b) for subchallenge 1, and Pearson correlation (c) and Spearman correlation (d) for subchallenge 2), performances shown for submissions are computed compound by compound and then averaged across compounds. The null hypothesis is generated for random predictions computed by random sampling, compound by compound, from the training set. (e,f) Performance of individual predictions (first boxplot, in red) is compared with performances of randomly aggregated predictions (wisdom of the crowds, in green) and with the aggregation of all predictions (last black bar). Performances are shown in terms of average Pearson correlation computed between predicted and measured values separately for each compound. Predictions were aggregated by averaging them. To aggregate only independent predictions, only one submission for each team was considered as the average of all predictions submitted by the team.
(a,b) Predictions were compared to the gold standard based on Pearson correlation for subchallenge 1 (a) and subchallenge 2 (b). The heatmap in a illustrates performances of all predictions for all compounds used for evaluation; predictions are ranked as in the final leaderboard and compounds are clustered. Pearson correlation values are saturated at −0.2 and 0.2. The heatmap in b illustrates performances of all ranked predictions for predicted median and interquantile range (q95–q05).
Prediction algorithms were also evaluated for their ability to categorically classify responses as cytotoxic or nontoxic using an EC10 threshold of 1.25 (as defined based on the classification of response curves described in Online Methods). 91 of 99 submissions achieved average AUC-ROC (area under the curve of a receiver operating characteristic) above 0.9, indicating that binary classification is much easier to predict than exact EC10 values.
We next assessed the contribution to prediction quality of RNA sequencing (RNA-seq) data, which were available for only a subset of cell lines. Overall, predictions were significantly better in the 97 cell lines with available RNA-seq data (Fig. 4, paired t-test, P < 10−16 for both r and pCi), which corresponds to a high effect size (Cohen's d equal to 1.6, derived from t-statistics). These results are consistent with a recent report that gene expression is more predictive of drug-induced cytotoxicity than genetic variation in cancer cell lines15.
(a,b) Performances of predictions for cell lines for which RNA-seq data were available were compared against performances of predictions for cell lines for which RNA-seq data were not available. Pearson correlation and pCi were computed for each compound; the comparison shows that predictions for cell lines for which RNA-seq data were available are significantly better (paired t-test, P << 10−10). All predictions are included in the analysis regardless of the actual use of the RNA-seq data.
Best performing method for subchallenge 1
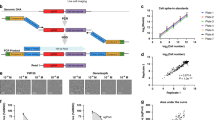
The best performing method for prediction of interindividual variation in cytotoxic response was able to predict with maximum r = 0.23 (average r = 0.05) and maximum pCi = 0.55 (average pCi = 0.51). As with the scoring analyses, this approach also omitted the 15 compounds that failed to induce a cytotoxic response from the analysis. Figure 5a shows the workflow of the prediction procedure for this method, which included steps for dimension reduction, prediction and cross-validation. A set of 0.15 M SNPs was selected for inclusion in this analysis using two approaches: (i) nonsynonymous SNPs within any gene as well as SNPs close to any gene defined by 2 kb upstream and 500 bp downstream regions; (ii) remaining SNPs if located within or close to gene members of the 41 KEGG20 gene sets (Supplementary Table 2) documented in the MSigDB database21 to represent cell cycle, cell death or cancer biology, or if they demonstrated correlation (P < 0.05) with the expression of at least one local gene based on the RNA-seq data (eQTL analysis). Information contained within this SNP set was then compiled into ten 'genetic clusters' using k-means clustering based on the first three principal components obtained by multidimensional scaling analysis22. The resultant 'genetic cluster' variable was highly representative of known geographic subpopulations (Fig. 5b), but also contained additional information not directly represented by each subpopulation. This variable was used to build a model of cytotoxicity for each compound using the Random Forest algorithm in conjunction with sex, geographic area and experimental batch. Cross-validation was carried out to choose parameters for clustering and to select methods for filtering SNPs. In the final scoring phase, this prediction approach achieved the best performance among dozens of submitted models, judged by the experimentally obtained true-response data. Details of this modeling approach are discussed further in Online Methods and Supplementary Predictive Models.
The prediction procedure of the best performing team of subchallenge 1. (a) Workflow of prediction for subchallenge 1. (b) Heatmap of number of cell lines in each category of “genetic cluster” (1–10, x axis) and geographic subpopulation (y axis). (c) Modeling workflow used by team QBRC for Toxicogenetics Challenge subchallenge 2. The model starts from deriving potential toxicity-related features by comparing response data and chemical descriptor profiles (step 1) and classifying compounds based on their toxicity responses (step 2). Then, group-specific models are built based on group-specific chemical features and the entire training set (step 3). Finally, the toxicity of a new compound is calculated as a weighted average of the predicted toxicities from each group-specific model (step 4). (d) In step 3, differentially distributed features and all training samples are used to develop group-specific models. (e) In step 4, model applicability domain and the similarities between the new compound and the compound group are used to determine the weights for each group-specific model.
Subchallenge 2: prediction of population-level parameters
Predictions of population-level parameters were scored using both Pearson correlation (r) and Spearman correlation (rs), with an approach similar to the previous subchallenge. For both statistics, the global performance of each submission was assessed by averaging correlations computed separately for median EC10 values, representing a 'typical' cytotoxicity response, and the difference between the 95th and 5th percentiles (interquantile range) for EC10 values, representing a measure of population dispersion.
The comparison with the null model of random predictions (Fig. 2c,d) was performed to assess the statistical significance of compound predictions. Of the 80 submitted predictions, the null hypothesis of randomly generated predictions was rejected (FDR < 0.05) for 13 predictions using average r and for 17 using average rs. For 13 predictions, the null hypothesis was rejected when both metrics were considered. A similar outcome was observed when using Fisher's method to assess the significance of individual submissions. Again, the ranking of best performing teams was shown to be robust with respect to the compounds used for scoring (Supplementary Fig. 4).
The average cytotoxicity (median EC10) of compounds appeared to be easier to predict (r ranged from −0.31 to 0.66; rs ranged from −0.29 to 0.72) than the variability in the response (interquantile range) of the population (r ranged from −0.22 to 0.37; rs ranged from −0.14 to 0.48; Supplementary Fig. 5).
Best performing method for subchallenge 2
The overall evaluation criterion for subchallenge 2 combined the prediction of median and interquantile range. The best performing method was able to predict the median and interquantile distance with r (and rs) equal to 0.52 (0.45) and 0.37 (0.40). The workflow used by this method (Fig. 5c), consisted of four major steps: feature selection, group identification, model development and test compound prediction. Features were selected from structural attributes of chemicals (step 1) derived in three ways and compared: CDK23 and SiRMS24 descriptors (both provided by the Challenge organizers) and Dragon descriptors25. Chemical descriptors were normalized separately and those descriptors that correlated with toxicity were used for training the models. The models using the Dragon descriptors achieved the best performance in both cross-validation and final scoring. In Step 2, compounds were categorized into four groups based on hierarchical clustering of their EC10 profiles across 487 cell lines. Random Forest models of cytotoxicity built separately for each compound group (Fig. 5c,d) were used to select features specific to prediction in that group, using all compounds to train the model. These models were used for predicting new compounds (Fig. 5c,e) as follows. For each new compound, toxicity was estimated using a weighted average of predictions from all four group-specific models, where weights were determined by similarity to each of the compound clusters. The similarity measure considered the distance to the cluster in the group-specific descriptor space, as well as the probability of being in the cluster using an additional classification model. The above modeling approaches were applied to predict both the median EC10 values and the interquantile range. For median EC10, cell-line specific predictions were generated using separate models and then averaged. For interquartile distance, a set of models was built to directly fit the measured interquantile distance for each compound. Further details of this modeling approach are discussed in Online Methods and Supplementary Predictive Models. Although this method was the best overall performer, other methods such as KSPA (see team Austria, Supplementary Predictive Models), provided more accurate prediction of the median cytotoxicity with r and rs equal to 0.65 and 0.72, respectively.
Predictability of compounds
Compounds were clearly separated into three clusters based on the accuracy of cytotoxicity predictions (Fig. 3a): a cluster of compounds for which predictions were high across all models (14 compounds), a cluster of compounds for which predictions were low across all models (17 compounds) and a cluster of compounds for which predictions varied across models. This separation was consistent between the two metrics used to evaluate performances (Supplementary Fig. 2). We next tested for features that could distinguish between compounds in the high-versus-low predictability clusters. Several chemical descriptors distinguished between high-versus-low predictability compounds and are listed in Supplementary Table 3. Notably, the Lipinski rule26 (i.e., a rule of thumb to evaluate drug similarity) was among these distinguishing features. As expected, compounds in the highly predictive cluster had lower pooled variance (thus are less noisy) than those in the poorly predicted cluster (one tailed t-test, P = 0.027). Contrary to expectation, compounds in the high-versus-low predictability clusters did not differ with regard to the distribution of cytotoxic response across the population in terms of median and interquantile range (Wilcoxon rank sum test, P = 0.65 and 0.68, respectively) nor to estimated heritability of compound cytotoxicities (P = 0.33; Supplementary Fig. 6). However, we observed that, when performing a principal component analysis on the cytotoxicity data (distributions centered to zero and scaled to unit variance for each compound), we could distinguish between the compounds with high and low predictability (Supplementary Fig. 7), indicating that the predictability was at least partially due to the cytotoxic profile of the compounds across the population.
Additionally, there was a clear difference in the shape of the distributions of the cytotoxic response across these two compound classes (Supplementary Fig. 8). In particular, highly predictable compounds tended to be characterized more frequently by a multimodal distribution (35% of highly predictable compounds, 0% of poorly predictable compounds, Hartigans's dip test for unimodality P < 0.05). The enrichment of multimodal distributions in highly predictable compounds suggests that algorithms are able to distinguish well when there are groups of individuals showing a different response to the same compound, rather than when the response follows a unimodal distribution. The principal component analysis allowed us also to estimate the predictive power that can be expected for new compounds. A linear support vector machine showed an accuracy of 66% (leave-one-out bootstrapping 5,000 times).
Wisdom of crowds
Previous DREAM challenges15,27,28 have observed that aggregation of predictions, which leverage the collective insight of all participants, can provide a more robust estimate than any individual prediction. We verified that, when applied to the test sets used for the challenge, the averaged aggregation across predictions within this study performed on par with top individual predictions for both subchallenge 1 (Supplementary Fig. 9a) and subchallenge 2 (Supplementary Fig. 9f). It is interesting to note that adding poor methods to an ensemble degrades the aggregate performance to a lesser degree than the gain resulting from adding good methods. For example, in subchallenge 1, the inclusion of the worst five individual predictions (whose correlations, from 0 to −0.015, had a range of 0.015) caused only a 5.55% decrease of performances based on Pearson correlation, whereas the inclusion of the top five individual predictions (whose correlations also had a range of 0.015, but from 0.03 to 0.045) caused a 21.17% increase (Supplementary Fig. 9d,e). The same trends have been observed in other analyses of ensemble methods27.
To robustly test whether aggregate model performance was consistently better at prediction relative to individual predictions, we next performed 200 iterations of an analysis in which optimized aggregate models were built using half of the test data and evaluated for performance using the remaining test data. Optimized aggregate models performed significantly better than the best individual prediction (paired t-test, P < 2.2 × 1016 for subchallenge 1 and 0.027 for subchallenge 2; Supplementary Fig. 9b,g). The optimal aggregate prediction outperformed the top individual prediction for all runs in subchallenge 1 and for 88.5% of the runs in subchallenge 2. Although optimized aggregate models built from the most accurate individual predictions have the best performance, in practice it is not possible to obtain an objective performance estimate for each model before analysis. For this reason, we also built an unsupervised aggregate model by combining a random selection of individual predictions. As a general trend, unsupervised aggregate models exhibited improved performance with respect to randomly selected individual predictions (Fig. 2e,f). Hence, as a general rule, it is preferable to aggregate efforts generated using different approaches even when the performance of the individual algorithms is unknown. Indeed, for both subchallenges 1 and 2, the aggregation of all predictions outperforms 87% of individual team predictions.
Characteristics of modeling approaches
We assessed modeling methodologies used within the challenge by surveying participants regarding their selection of input data, preprocessing methodologies for data reduction, prediction algorithms and techniques for model validation (Fig. 6). All predictions used at least one of the data sources provided by the organizers. Notably, some models performed well using only the sex/ancestry covariates in the absence of genomic data (subchallenge 1, Team ranked 4th). Most of the teams, including the best performing teams, also integrated information from external sources into their models (24% for subchallenge 1 and 47% for subchallenge 2). A variety of methodological algorithms representing the state of the-art in the modeling field were applied in both subchallenges. No methodologies for data reduction, predictive modeling or model validation used in this challenge outperformed the others in any obvious manner (Supplementary Fig. 10), suggesting that performance was dependent mainly on strategy for methodological application rather than on algorithmic choice.
Overview of the input data, data reduction techniques, prediction algorithms and model validation techniques used by participants to solve the challenge. Participants were asked to fill out a survey in order to be included in this publication as part of the NIEHS-NCATS-UNC Dream Toxicogenetics challenge consortium; only data for teams that filled out the survey are shown here. Each row corresponds to a submission, and they are ordered based on the final rank for subchallenge 1 and subchallenge 2, respectively. Data originate from 75 filled-out surveys for subchallenge 1 (of 99 submissions) and 51 filled-out surveys for subchallenge 2 (of 80 submissions). This corresponds to 21 (of 34) teams for subchallenge 1 and 12 (of 23) for subchallenge 2.
Discussion
The results of the NIEHS-NCATS-UNC-DREAM Toxicogenetics Challenge demonstrate that modeling algorithms were able to predict cytotoxicity traits based on genetic profiles with higher than random accuracy, although results were modest. The methods developed for subchallenge 1 are likely to be even more useful in future settings, as decreasing sequencing costs means that larger training sets will be available to achieve higher predictive accuracy.
Accurate predictions of population-level cytotoxicity could help to establish safe environmental exposure limits. Therefore, we tested whether population-level cytotoxicity could be predicted based on the structural attributes of compounds (subchallenge 2). Participants were able to robustly predict both mean toxicity and population variability in cytotoxicity based entirely on chemical attributes of compounds. These results demonstrate that predictive algorithms may be able to provide real-world benefit in environmental risk assessment and suggest an opportunity to incorporate structural predictions into hazard assessments of new compounds.
The ability to predict interindividual variability in cytotoxic response (subchallenge 1) is consistent with predictive performances for complex genetic traits such as height29. Comparable predictive performances were also observed in a recent DREAM analysis comparing algorithms that predicted cellular response to cancer agents15. Because each individual SNP describes only a small portion of overall variation in response, the ability to accurately predict phenotype, or even to detect true genetic signal in large-scale genomic analysis, requires very large sample sizes30,31. Downsampling analyses suggest that predictability would increase with additional samples (Supplementary Fig. 11).
The complex nature of the EC10 cytotoxic phenotype, which is a statistic that can be influenced by often high levels of technical variation9, may also decrease prediction power. Indeed, we observed that algorithms were able to broadly classify compounds as cytotoxic or noncytotoxic more accurately than they could predict cytotoxicity. We also observed that cytotoxic predictability varied across compounds although we could not identify a clear pattern as to which characteristics improve predictability.
Regardless of these limitations, prediction rules identified within best performing algorithms can be leveraged to advance future efforts within this field regarding data collection and prediction analysis. In particular, the best performing team in subchallenge 1 developed a novel data processing approach that incorporated prior biological knowledge into their machine-learning methodology by clustering individual cell lines based on variants located in a set of presumed biologically relevant loci. The resultant clusters were broadly representative of geographically distinct subpopulations but included additional information. This approach may be generally useful in predicting complex traits based on genetic variant data. The prediction approaches developed for subchallenge 2 could be generalized to predict and rationalize chemical compounds' biological properties from chemical structures. The proposed model incorporated the population-level structures in toxicity profiles into traditional, quantitative, structure-activity relationship models.
A unique aspect of this study is that it focuses on the use of constitutional genetic variation to predict toxicity response. Although some studies support the idea that transcriptomic profiles can provide higher predictive performance than genetic profiles—and, indeed, we observed improved predictive performance with the availability of baseline transcriptional data—the use of transcriptional data has often been upon perturbation, which is not applicable within our framework of environmental risk assessment. As such, the use of genetic profiles to predict variation in response—across individuals and across compounds—on the population level provides a tool that can be applied within real-world applications.
Overall, our analyses assessed the capability of computational approaches to provide meaningful predictions of cytotoxic response to environmental compound exposure using genetic and chemical structure information. The models developed within this project would require higher accuracy to provide actionable information at the level of an individual. However, for prioritization of compounds, this study provides statistically significant evidence for the ability to predict toxic effects on populations using a stringent evaluation methodology.
Methods
Data description.
A schematic of the challenge is outlined in Figure 1. The cytotoxicity data used in the challenge consist of the estimated effective concentrations that induced a 10% decrease in viability (i.e., the EC10) generated for 884 lymphoblastoid cell lines in response to 156 common environmental compounds. Participants were provided with a training set of cytotoxicity data for 620 cell lines and 106 compounds along with genotype data for all cell lines, RNA-seq data for 337 cell lines and chemical attributes for all compounds. Primary data generation and other details on the cytotoxicity screening are detailed in Abdo et al.5 available in open access at: http://ehp.niehs.nih.gov/1408775/. A brief description of the data is provided below. A total of four toxicity phenotyping, genomic, genotyping and chemical attribute data sets were available for this challenge. Descriptions of each data set can be found in the annotation files associated with each data set supplied through the DREAM website (https://www.synapse.org/#!Synapse:syn1761567/wiki/56224).
(1) Chemicals. Chemicals were a subset of the National Toxicology Program's 1408 chemical library as detailed in Xia et al.32 and were selected to broadly represent chemicals found in the environment and consumer products. A small number of pharmaceuticals was included as well, but the focus of this experiment was on environmental toxicants. Chemicals were dissolved in dimethyl sulfoxide (DMSO) to prepare 20 mM stock concentrations, and then diluted in DMSO to provide final concentrations ranging from 0.33 nM to 92 μM. We fit all chemicals and concentrations, including duplicate samples for 8 chemicals, and positive and negative controls for each cell line to a single 1,536-well plate.
(2) Cell lines. We acquired the immortalized lymphoblastoid cell lines from Coriell. We aimed to represent human genetic diversity and thus selected cell lines from nine populations across the globe from Europe, the Americas, Asia and Africa. These included the Han Chinese in Beijing, China (CHB); Japanese in Tokyo, Japan (JPT); Luhya in Webuye, Kenya (LWK); Yoruban in Ibadan, Nigeria (YRI); Utah residents with European ancestry (CEU); British from England and Scotland (GBR); Tuscans in Italy (TSI); Mexican ancestry in Los Angeles, California (MXL); and Colombians in Medellín, Colombia (CLM). Cell lines were selected to reflect unrelated individuals by removing all instances of first-degree relatives. Within each population, cell lines were included from both males and females. Cell lines were randomly divided into five screening batches with equal distribution of populations and gender. Approximately 65% of the cell lines were seeded for repeat analysis on multiple plates (2–3 plates per batch and/or between batches).
(3) Cytotoxicity profiling. Screening was performed in 1,536-well plate format. The negative control was DMSO at 0.46% vol/vol; the positive control was tetra-octyl-ammonium bromide (46 μM). We used the CellTiter-Glo Luminescent Cell Viability assay (Promega) to assess intracellular ATP concentration, a marker for viability/cytotoxicity, at 40 h post treatment. We used a ViewLux plate reader (PerkinElmer) to detect luminescent intensity. Cytotoxicity data were divided into two parts for training and external validation of the models. The former contained individual EC10 values in a 487 cell line × 106 compound matrix. The latter contained population-level summary statistics of EC10 values per compound in a 106 compound × 3 statistics matrix. EC10 values were calculated from concentration-response exposure data for each chemical across all 884 cell lines and were normalized relative to the positive/negative controls. The EC10 value is defined as the concentration at which intracelluar ATP content was decreased by 10% and was estimated for each cell line by normalizing data to vehicle-treated cells and then fitting normalized concentration-response curves to a three parameter logistic regression model where maximum response was fixed to −100% and minimum response was derived from the response of the lowest three concentrations, with the exclusion of outliers as defined by >2 s.d. If the compound had less than 10% effect over the range of concentrations used in the experiment, the EC10 was set to 100 μM to represent a “no observable adverse effect level.” EC10 values were batch corrected using Combat33 and then replicate values per individual were averaged. Batch information is provided in the covariate file, although data have already been corrected for this technical source of variation. Data providers verified that EC10 and EC50 values are reasonably correlated. However, only EC10 values were used for the challenge, as this is the most interesting measure for low-dose toxic response and highest relevance for susceptible subpopulations.
(4) Genotype data. Approximately 1.3 million SNPs are provided for each individual. For 761 cell lines, these SNPs were directly genotyped using the Illumina HumanOmni2.5 platform. For the remaining 123 unrelated individuals, the available sequencing data were used to impute the missing SNPs, using either HapMap3 genotypes for these individuals, or sequence data, using MACH software and the reference set for imputation. SNPs with a call rate below 95%, MAF < 0.01, strong evidence against Hardy-Weinberg disequilibrium (P < 10−6) were excluded from this data set, leaving a final set of 1,327,016 SNPs.
(5) RNA sequencing. RNA sequencing data were available for 337 of the cell lines (representing the CEU, GBR, TSI and YRI subpopulations) as data set E-GEUV-1 in the ArrayExpress repository. The mapped reads files (BAM format) were downloaded and IsoDOT was used to count the reads of each nonoverlapping exon, which has been preprocessed in IsoDOT library files. Read counts for each gene were generated by summing the read counts for all of that gene's exons. Data are provided as raw gene counts for 46,256 transcripts.
(6) Chemical attributes. Each compound is described by quantitative structural attributes as developed using two standard methodologies for the purpose of providing a standardized description of structural properties that are common across chemicals and can be used to model structure-based commonalities in cytotoxicity. Attributes include 160 chemical descriptors calculated using the Chemistry Development Kit (CDK)23. In addition, 9,272 chemical descriptors were generated for each compound using the Simplex representation of molecular structure (SIRMS)24. In this process, each molecule is represented as a system of tetratomic fragments with fixed composition, structure, chirality and symmetry as described here. Data were not further processed or normalized.
Web-based resource and challenge rules.
The challenge was hosted on Synapse34, a cloud-based platform for collaborative scientific data analysis. Synapse was used to distribute challenge data and to track participant agreement to the appropriate data usage conditions (main challenge web page https://www.synapse.org/#!Synapse:syn1761567). Synapse was used also to run a real-time leaderboard during the first phase of the challenge, where participant could submit their prediction on a test set (133 cell lines) that was then released as part of the training set for the final phase of the challenge, and receive real time feedback on their performances (Supplementary Fig. 12).
For the final submissions, challenge participants created Synapse projects (https://www.synapse.org/#!Synapse:syn1840307/wiki/67255) containing their predictions (maximum five predictions per team) together with the code used to derive them and wikis in which participants describe their methods in prose and figures. To assure reproducibility of the challenge, the organizers of submissions ran the code of the best performing methods. All data are stored in synapse and are available in Synapse (https://www.synapse.org/#!Synapse:syn1761567/wiki/56224).
Supplementary Figures, Supplementary Tables, Supplementary Predictive Models and Supplementary Code are is also available in Synapse as an interactive resource https://www.synapse.org/#!Synapse:syn1840307.
Software and statistical methods.
R (v3.1) was used for scoring and for post-challenge analysis and False Discovery Rate (FDR) was computed using the Benjamini–Hochberg procedure. R (v2.15) and plink (v1.07) were used by the best performing team of subchallenge 1. R (v2.15) and Dragon (v5.5) were used by the best performing team of subchallenge 2. All relevant code has been provided as Supplementary Code and can also be found online (https://www.synapse.org/#!Synapse:syn1840307/wiki/231104). The file includes: a) scoring functions, b) code used to generate supplementary figures, c) code submitted by participants and used to generate predictions.
Selection of training and test set.
For both subchallenges, the data set was divided in training and test set using stratified random sampling to guarantee that extreme responses were included in both training and test data. More in details, for subchallenge 1, the data set was clustered in N groups based on EC10 profiles, where N is the number of individuals in the test set. One individual is then selected from each group to be part of the test set. This guarantees that strong and weak responses were present in both the training and the test sets, which is also more representative of the task of predictive genomics. The same approach was used for subchallenge 2, but with clustering by compounds instead of by individuals.
Scoring algorithm for subchallenge 1.
For each metric (i.e., Pearson correlation and prob C-index), teams were ranked separately for each compound and then the average rank was computed, for each team, across compounds, providing a final rank for each metric. Teams were finally ranked (final rank) based on the average of the rank computed for each metric (mean ranking). Only 91 out of 106 compounds were used for final scoring as 15 compounds were shown to have no toxicity across the human cell population (Supplementary Fig. 1a). Since the aim of the challenge is the prediction of how individual cell lines differently respond to each compound, compounds for which the response is the same across all the population are not interesting in this context and were therefore excluded from the evaluation to avoid including noise in the scoring metric.
Significance and robustness of subchallenge 1.
Significance of prediction for individual compounds. By comparing the distribution of submissions with respect to the null model of randomly predicted EC10 values, we verified that predictions are significantly better then random for 55 of 91 compounds (Wilcoxon sum rank test, P < 0.05) if we consider r and pCi separately, and for 59 of 91 compounds if we consider compounds for which the hypothesis of equal distribution is rejected for at least one metric. To assess whether lack of predictability was universal across submissions, we repeated this analysis within the subset of 25 submissions with the best prediction for each compound. In this case, the alternative hypothesis is accepted for all 91 compounds for both r and pCi.
Significance of predictions based on ranking. The mean ranking across compounds was compared with the empirical null distribution of 100,000 randomized mean rankings, derived by randomly ranking teams for each compound and then computing the randomized mean ranking (Supplementary Table 4). Of the 99 predictions, the null model could be rejected (t-test, FDR < 0.05) for 17 predictions, considering the mean ranking computed based on Pearson correlation, and for 17 predictions, when considering pCi. For 15 predictions, the null model was rejected considering both metrics.
Robustness analysis. To assess the robustness of the final team rank with respect to the compounds used for scoring, we recomputed the score multiple times by randomly masking each time, data for 10% of the compounds. In Supplementary Figure 3, we compared the distribution of the mean ranking and of the final rank obtained by all teams and verified that the best submission is reliably ranked first as it is significantly on top with respect to all the other submissions (one-sided Wilcoxon signed-rank test, FDR < 10−10). The robustness analysis also shows that all of the top six submissions are statistically different.
Classification problem.
Considering the inherent variances in the measured EC10 values, the model performances were reevaluated for their significance in predicting the activity outcome of a compound in a cell line (i.e., cytotoxic or nontoxic), instead of the exact EC10 value. As shown in Supplementary Figure 13a, most of the compounds in the test set were either active/cytotoxic (43%) in all of the cell lines or inactive/nontoxic (16%) across all of the cell lines, whereas each cell line showed a well-balanced number of active/inactive calls. To evaluate overall model performance at classifying active and inactive compounds, for each model submission, AUC-ROC values were calculated first for each cell line then averaged across all cell lines (Supplementary Fig. 13b, red line). With a few exceptions, most model submissions (91 out of 99) performed well with average AUC-ROC values >0.9. To further assess model performance, model sensitivity (recall) was calculated for compounds that were active across all cell lines to test the model's capacity at correctly making active calls, and specificity was calculated for compounds that were inactive across all cell lines to test the model's capacity at correctly making inactive calls. For this analysis, the active EC10 cutoff was determined by comparing the experimental EC10 values of all test set compounds to their cytotoxic/nontoxic classifications. An EC10 of 1.25 μM was chosen as the optimal cutoff for classifying a compound as active in a cell line. The average sensitivity and specificity measures calculated for all model submissions are shown in Supplementary Figure 13b. Most model submissions achieved good sensitivity (Supplementary Fig. 13b; green line) in predicting active compounds and good specificity (Supplementary Fig. 13b; purple line) in predicting inactive compounds with 84 out of the 99 submissions achieving >90% average sensitivity and 91 submissions achieving >90% average specificity.
Evaluation of model performance. Compounds were assigned one of the four categories, true positive (TP), false positive (FP), true negative (TN) and false negative (FN), based on their activity observed in the assay and model predicted activity according to the following scheme.

The numbers of TP, FP, TN and FN calls were counted at various EC10 cutoffs and the AUC-ROC was calculated for each compound in each cell line. The ROC curve is a plot of sensitivity against 1-specificity, where sensitivity is defined as TP/(TP+FN) and specificity defined as TN/(FP+TN). A perfect model would have an AUC-ROC of 1 and an AUC-ROC of 0.5 or lower indicates that the model predictions are not better than random.
Best performing method for subchallenge 1.
We evaluated the effect of removing any one of the four predictors used in the random Forest algorithm on the prediction. Supplementary Figure 14 shows that removing any of the four predictors leads to a worse prediction accuracy measured by Pearson correlation. We also investigated whether the filtering step with KEGG pathway and RNA-seq data gives any improvement in prediction accuracy by randomly sampling 0.15 million SNPs from the 0.61 million SNPs selected from the first step of feature selection or directly using the 0.61 million SNPs to generate principal components. Indeed, Supplementary Figure 14 shows that both methods lead to a mean Pearson correlation that is even smaller than a prediction model that does not include the “genetic cluster” variable. In conclusion, our results suggest that the four variables (sex, population, experimental batch and “genetic cluster”) all contribute to the prediction accuracy and that the second round of filtering with KEGG pathway and RNA-seq data helps to generate a “genetic cluster” variable that carries meaningful information regarding compound toxicity.
Scoring algorithm for subchallenge 2.
Predictions were ranked separately for each metric (i.e., Pearson correlation and Spearman correlation) by computing the average rank of each team for the predicted median and interquantile distance. The final rank was thus computed based on the average of the rank computed using the two metrics (mean ranking).
Significance and robustness of subchallenge 2.
Significance of predictions based on Fisher's method. The statistical significance of predictions was verified by combining, using Fisher's method, the P-value computed separately for the performances of each submission in predicting the median and the interquantile distance. Performances are above what is expected at random (FDR < 0.05) for 26 submissions when considering Pearson correlation, for 39 submissions when considering Spearman correlation, and for 24 submissions when considering both metrics (Supplementary Table 5).
Robustness analysis. The robustness analysis (Supplementary Fig. 4), computed as described for subchallenge 1, showed that the best performing team is robustly ranked first with all five submissions outperforming submissions from other teams (one-sided Wilcoxon signed-rank test, FDR < 10−10). As shown from the FDR analysis, the top two submissions are not statistically distinguishable.
Best performing method for subchallenge 2.
To predict chemical toxicities from chemical profiles, we developed computational models with four steps. The first step is feature selection. The curated chemical structure (provided by the organizer in Structure Data Format (SDF)) was used to generate Dragon25 descriptors for each compound. The derived descriptor matrices were range scaled to 0∼1, and those with low variance (s.d. < 10−6) were excluded. For any pair of highly correlated descriptors (Pearson correlation, P > 0.95), one descriptor was removed randomly. The descriptors were then filtered based on their correlation to compounds' cytotoxicity (EC10 values). There are 67 descriptors that are significantly (Pearson correlation, P < 0.05) correlated to EC10 values in >70% of the cell lines.
The second step is to evaluate the toxicity distributions of the compounds, and to determine the compound groups based on their toxicity profiles. We divided the 106 compounds into four groups based on hierarchical clustering of their EC10 profiles across 487 cell lines.
The third step is to develop group-specific models (Fig. 5c). For each group identified in step 2, we used ANOVA to select features that are specific for compounds in the group versus compounds in the remaining groups. Then, the values of the selected features for all training compounds (91 compounds with measurable toxicity values) were used as the training data. Therefore, there are four Random Forest models that are specific to each cluster of compounds (model M1, M2, M3, M4).
The final step is to apply the models for predicting new compounds (Fig. 5d). For each new compound, we estimated its toxicity by a weighted average of its predictions from all four group-specific models. The weights were determined by its similarity to each of the compound clusters. The similarity involves calculating the compound's distance to the cluster in the group-specific descriptor space, as well as its probability of being in the cluster using a classification model. If the compound's distance to one cluster is smaller than a distance threshold, we think that the cluster-specific model is appropriate to predict the new compound, and the weight is proportional to its probability of being in that group. Otherwise, we think the model is inappropriate to predict the new compound, and its weight for predicting this specific compound would be 0. The distance threshold is determined by the applicability domain described by Zhen and Tropsha35 (with parameter z = 2). In certain cases, where the new compound is out of the threshold for all four group-specific models, we predicted its activity using the entire training set and all the descriptors.
We apply the above modeling approaches to estimate both the median EC10 values and the interquartile distance for new compounds, but with small modifications. To predict a compounds' median EC10 value, we built separate models to fit EC10 values measured from individual cell lines. We then derived the median EC10 value from the predicted cell-line-specific EC10 values. To predict a compounds' interquartile distance, we built a single set of models to fit the measured interquartile distance directly.
Predictability of compounds.
We compared the cytotoxic response of all individuals to the two groups of compounds as shown in Supplementary Figure 8; in this case the null hypothesis of equal mean of the two groups is now rejected (P < 2.2 × 10−16) and, notably, there is a clear difference in the shape of the two distributions. The first possible reason for the bimodal distribution of EC10 values of highly predictable compounds is that this group of compounds show very high or very low toxicity in all of the population; however, this is not the case because, if we test for multimodality the median EC10 values of highly predictable compounds, we verify that the distribution is unimodal (Hartigans's dip test, P = 0.70). The second possible reason is that highly predictable compounds are the ones that show multimodal distribution across the population; applying the Hartigans's dip test for unmodality on all compounds (distribution of EC10 values across individuals), we verify that 35.71% (5 of 14) of the highly predictable compounds have a multimodal distribution (P < 0.05, Supplementary Fig. 8b for two examples), whereas 0% (0 of 17) of the poorly predictable compounds have a multimodal distribution (they all have unimodal distribution; Supplementary Figure 8c for two examples). The percentage of multimodal distributions is 10% for the remaining compounds (6 of 60).
Noise in the data. The distribution of the pooled variance for compounds with high predictability is slightly but significantly shifted to the left with respect to the distribution for compounds with low predictability (one tailed t-test, P = 0.027). Thus, as expected, noisy compounds are in general harder to predict with respect to compounds with a lower pooled variance.
Survey data analysis.
We received responses for 75 submissions (out of 99) for subchallenge 1 and for 51 submissions (out of 80) for subchallenge 2. This corresponds to 21 (out of 34) teams for subchallenge 1, and 14 (out of 23) for subchallenge 2. An overview of the information provided by the survey is shown in Figure 6. The effect of used data and methods on the performances of submissions is shown in Supplementary Figure 10 in terms of average Pearson correlation. To deal with the fact that each team submitted up to five submissions that might not be independent of each other, predictions using the same data and methods (based on the information from the survey) were averaged and considered as one prediction. Using this approach, we obtained 49 independent submissions for subchallenge 1 and 28 for subchallenge 2. Data and methods listed as “others” in Supplementary Figure 10 and Figure 6 are reported in Supplementary Table 6.
Input data used for predictions. To solve subchallenge 1, 89% of the participants who replied to the survey, used the SNPs data provided by the organizers either alone or along with other data using additional sources (e.g., pathway information, GO terms) to filter them. RNA-seq data were used for almost half (47%) of the submissions and this was shown to provide an overall improvement of performances. Only a minority of the participants (16%) also included in their predictive model information about chemical descriptors.
For subchallenge 2, most submissions (78%) did not take into account any genetic information to predict the cytotoxicity of new compounds. As for the chemical features, about 76% made use of at least one of the chemical descriptors provided by the organizers (CDK and SiRMS), but many teams (45%) included in addition or exclusively information from other sources like ChEMBL36 and PubChem37 public databases or different chemical descriptors like Dragon25 or ECFP38 (see Supplementary Table 6 for the full list).
Accession codes.
All data used for the challenge are available in Synapse (syn1761567).
Change history
01 October 2015
In the version of this article initially published, in the HTML only, all authors names were incorrectly included in the main author list, and several authors names were repeated. The authors have now added 12 additional authors to the list of “Other participants in the NIEHS-NCATS-UNC DREAM Toxicogenetics Collaboration,” including Alok Jaiswal, Antti Poso, Himanshu Chheda, Ismeet Kaur, Jing Tang, John-Patrick Mpindi, Krister Wennerberg, Natalio Krasnogor, Samuel Kaski, Tero Aittokallio, Petteri Hintsanen and Suleiman Ali Khan. Names in this list that were also in the list of “top-performing teams” have been deleted. In addition, affiliation number 3 (Univ. Texas) has been added to Xiaowei Zhang’s name in the main author list. The errors have been corrected in the HTML and PDF versions of the article.
References
Judson, R. et al. The toxicity data landscape for environmental chemicals. Environ. Health Perspect. 117, 685–695 (2009).
Jacobs, A.C. & Hatfield, K.P. History of chronic toxicity and animal carcinogenicity studies for pharmaceuticals. Vet. Pathol. 50, 324–333 (2013).
Zeise, L. et al. Addressing human variability in next-generation human health risk assessments of environmental chemicals. Environ. Health Perspect. 121, 23–31 (2013).
Dorne, J.L.C.M. Metabolism, variability and risk assessment. Toxicology 268, 156–164 (2010).
Abdo, N. et al. Population-based in vitro hazard and concentration-response assessment of chemicals: the 1000 Genomes high-throughput screening Study. Environ. Health Perspect. 123, 458–466 (2015).
Burczynski, M.E. et al. Toxicogenomics-based discrimination of toxic mechanism in HepG2 human hepatoma cells. Toxicol. Sci. 58, 399–415 (2000).
Uehara, T. et al. Prediction model of potential hepatocarcinogenicity of rat hepatocarcinogens using a large-scale toxicogenomics database. Toxicol. Appl. Pharmacol. 255, 297–306 (2011).
Kleinstreuer, N.C. et al. Phenotypic screening of the ToxCast chemical library to classify toxic and therapeutic mechanisms. Nat. Biotechnol. 32, 583–591 (2014).
Choy, E. et al. Genetic analysis of human traits in vitro: drug response and gene expression in lymphoblastoid cell lines. PLoS Genet. 4, e1000287 (2008).
Caliskan, M., Cusanovich, D.A., Ober, C. & Gilad, Y. The effects of EBV transformation on gene expression levels and methylation profiles. Hum. Mol. Genet. 20, 1643–1652 (2011).
Mangravite, L.M. et al. A statin-dependent QTL for GATM expression is associated with statin-induced myopathy. Nature 502, 377–380 (2013).
Gamazon, E.R. et al. Comprehensive genetic analysis of cytarabine sensitivity in a cell-based model identifies polymorphisms associated with outcome in AML patients. Blood 121, 4366–4376 (2013).
Collins, F.S., Gray, G.M. & Bucher, J.R. Toxicology: transforming environmental health protection. Science 319, 906–907 (2008).
Margolin, A.A. et al. Systematic analysis of challenge-driven improvements in molecular prognostic models for breast cancer. Sci. Transl. Med. 5, 181re1 (2013).
Costello, J.C. et al. A community effort to assess and improve drug sensitivity prediction algorithms. Nat. Biotechnol. 32, 1202–1212 (2014).
1000 Genomes Project Consortium. et al. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012).
1000 Genomes Project Consortium. et al. A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073 (2010).
Lappalainen, T. et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature 501, 506–511 (2013).
Brown, C.C. et al. Genome-wide association and pharmacological profiling of 29 anticancer agents using lymphoblastoid cell lines. Pharmacogenomics 15, 137–146 (2014).
Kanehisa, M. et al. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 42, D199–D205 (2014).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Steinbeck, C. et al. The Chemistry Development Kit (CDK): an open-source java library for chemo- and bioinformatics. J. Chem. Inf. Comput. Sci. 43, 493–500 (2003).
Kuz'min, V.E., Artemenko, A.G. & Muratov, E.N. Hierarchical QSAR technology based on the Simplex representation of molecular structure. J. Comput. Aided Mol. Des. 22, 403–421 (2008).
Todeschini, R., Consonni, V., Mauri, A. & Pavan, M. DRAGON-software for the calculation of molecular descriptors. Web version 3 (2004).
Lipinski, C.A., Lombardo, F., Dominy, B.W. & Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 46, 3–26 (2001).
Marbach, D. et al. Wisdom of crowds for robust gene network inference. Nat. Methods 9, 796–804 (2012).
Meyer, P. et al. Network topology and parameter estimation: from experimental design methods to gene regulatory network kinetics using a community based approach. BMC Syst. Biol. 8, 13 (2014).
Yang, J. et al. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 42, 565–569 (2010).
Park, J.-H. et al. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat. Genet. 42, 570–575 (2010).
Chatterjee, N. et al. Projecting the performance of risk prediction based on polygenic analyses of genome-wide association studies. Nat. Genet. 45, 400–405 (2013).
Xia, M. et al. Compound cytotoxicity profiling using quantitative high-throughput screening. Environ. Health Perspect. 116, 284–291 (2008).
Johnson, W.E., Li, C. & Rabinovic, A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8, 118–127 (2007).
Derry, J.M.J. et al. Developing predictive molecular maps of human disease through community-based modeling. Nat. Genet. 44, 127–130 (2012).
Zheng, W. & Tropsha, A. Novel Variable Selection Quantitative Structure-Property Relationship Approach Based on the k-Nearest-Neighbor Principle. J. Chem. Inf. Comput. Sci. 40, 185–194 (2000).
Gaulton, A. et al. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 40, D1100–D1107 (2012).
Wang, Y. et al. PubChem's BioAssay Database. Nucleic Acids Res. 40, D400–D412 (2012).
Rogers, D. & Hahn, M. Extended-connectivity fingerprints. J. Chem. Inf. Model. 50, 742–754 (2010).
Acknowledgements
This research was supported in part by the US National Institutes of Health (NIH), National Institute of Environmental Health Sciences (NIEHS). This work was made possible by US Environmental Protection Agency grants STAR RD83516601 and RD83382501, NIH grants R01CA161608 and R01HG006292, and through an interagency agreement (IAG #Y2-ES-7020-01) from NIEHS to NCATS. F.E. thanks European Molecular Biology Laboratory Interdisciplinary Post-Docs (EMBL EIPOD) and Marie Curie Actions (COFUND) for funding. Best performing team was funded by NIH grants 5R01CA152301 and 1R01CA172211.
Author information
Authors and Affiliations
Consortia
Contributions
F.E. designed the analyses, scored predictions, performed computational analyses of challenge outcomes and wrote the manuscript. L.M.M. led project design and implementation including data collection from participants and participated in data analysis and manuscript development. J.C.B. and M.K. implemented the leaderboard and final scoring of predictions, collection of code, methods and outcomes, and participated in writing the manuscript supplement. T.N. and S.F. participated in project design and development. A.D., R.T., R.H., M.X., A.S., N.A., O.K., I.R. and F.A.W. generated and processed the data, and contributed to project design and interpretation of results. I.R., F.A.W. and R.T. participated in writing the manuscript. M.P.M. contributed to development and implementation of methodologies to score predictions. T.W., H.T., X.Z., J.Y., R.Z., G.X. and Y.X. (led by T.W., H.T. and Y.X.) participated in the challenge as modelers, developing the model with the best predictive performance, participated in analysis of challenge outcomes and participated in writing the manuscript. J.S.-R. and G.S. were responsible for overall design, development and management of project and participated in writing the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Challenge organizers: Federica Eduati, Lara M Mangravite, J Christopher Bare, Thea Norman, Mike Kellen, Michael P Menden, Stephen Friend, Gustavo Stolovitzky & Julio Saez-Rodriguez Data producers: NIEHS: Allen Dearry & Raymond R Tice NCATS: Ruili Huang, Menghang Xia & Anton Simeonov UNC: Nour Abdo, Oksana Kosyk, Ivan Rusyn & Fred A Wright Top-performing teams: Subchallenge 1: Tao Wang, Hao Tang, Xiaowei Zhan, Jichen Yang, Rui Zhong, Guanghua Xiao & Yang Xie Subchallenge 2: Hao Tang, Jichen Yang, Tao Wang, Guanghua Xiao & Yang Xie Other participants in the NIEHS-NCATS-UNC DREAM Toxicogenetics Collaboration: Tero Aittokallio, Salvatore Alaimo, Alicia Amadoz, Muhammad Ammad-ud-din, Chloé-Agathe Azencott, Jaume Bacardit, Pelham Barron, Elsa Bernard, Andreas Beyer, Shao Bin, Alena van Bömmel, Karsten Borgwardt, April M Brys, Brian Caffrey, Jeffrey Chang, Jungsoo Chang, Himanshu Chheda, Eleni G Christodoulou, Mathieu Clément-Ziza, Trevor Cohen, Marianne Cowherd, Sofie Demeyer, Joaquin Dopazo, Joel D Elhard, Andre O Falcao, Alfredo Ferro, David A Friedenberg, Rosalba Giugno, Yunguo Gong, Jenni W Gorospe, Courtney A Granville, Dominik Grimm, Matthias Heinig, Rosa D Hernansaiz, Petteri Hintsanen, Sepp Hochreiter, Liang-Chin Huang, Matthew Huska, Alok Jaiswal, Yunlong Jiao, Samuel Kaski, Ismeet Kaur, Suleiman Ali Khan, Günter Klambauer, Natalio Krasnogor, Michael Kuhn, Miron Bartosz Kursa, Rintu Kutum, Nicola Lazzarini, Inhan Lee, Michael K K Leung, Weng Khong Lim, Charlie Liu, Felipe Llinares López, Alessandro Mammana, Andreas Mayr, Tom Michoel, Misael Mongioví, Jonathan D Moore, John-Patrick Mpindi, Ravi Narasimhan, Stephen O Opiyo, Gaurav Pandey, Andrea L Peabody, Juliane Perner, Antti Poso, Alfredo Pulvirenti, Konrad Rawlik, Susanne Reinhardt, Carol G Riffle, Douglas Ruderfer, Aaron J Sander, Richard S Savage, Erwan Scornet, Patricia Sebastian-Leon, Roded Sharan, Carl Johann Simon-Gabriel, Veronique Stoven, Jingchun Sun, Jing Tang, Ana L Teixeira, Albert Tenesa, Jean-Philippe Vert, Martin Vingron, Thomas Walter, Krister Wennerberg, Sean Whalen, Zofia Wiśniewska, Yonghui Wu, Hua Xu, Shihua Zhang, Junfei Zhao, W Jim Zheng, Dai Ziwei
Joint Research Center for Computational Biomedicine (JRC-COMBINE), RWTH Aachen University, Aachen, Germany.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–14 (PDF 2771 kb)
Supplementary Tables
Supplementary Tables 1–7 (PDF 1196 kb)
Supplementary Information
Predictive Models (PDF 447 kb)
Rights and permissions
This work is licensed under a Creative Commons Attribution- NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/.
About this article
Cite this article
Eduati, F., Mangravite, L., Wang, T. et al. Prediction of human population responses to toxic compounds by a collaborative competition. Nat Biotechnol 33, 933–940 (2015). https://doi.org/10.1038/nbt.3299
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.3299
This article is cited by
-
Simplex representation of molecular structure as universal QSAR/QSPR tool
Structural Chemistry (2021)
-
QSAR-derived affinity fingerprints (part 2): modeling performance for potency prediction
Journal of Cheminformatics (2020)
-
Toxicity testing in the 21st century: progress in the past decade and future perspectives
Archives of Toxicology (2020)
-
DRAP: a toolbox for drug response analysis and visualization tailored for preclinical drug testing on patient-derived xenograft models
Journal of Translational Medicine (2019)
-
A microfluidics platform for combinatorial drug screening on cancer biopsies
Nature Communications (2018)