Abstract
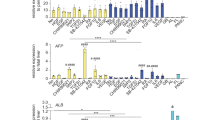
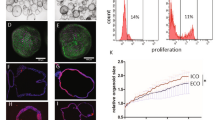
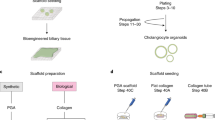
Although bile duct disorders are well-recognized causes of liver disease, the molecular and cellular events leading to biliary dysfunction are poorly understood. To enable modeling and drug discovery for biliary disease, we describe a protocol that achieves efficient differentiation of biliary epithelial cells (cholangiocytes) from human pluripotent stem cells (hPSCs) through delivery of developmentally relevant cues, including NOTCH signaling. Using three-dimensional culture, the protocol yields cystic and/or ductal structures that express mature biliary markers, including apical sodium-dependent bile acid transporter, secretin receptor, cilia and cystic fibrosis transmembrane conductance regulator (CFTR). We demonstrate that hPSC-derived cholangiocytes possess epithelial functions, including rhodamine efflux and CFTR-mediated fluid secretion. Furthermore, we show that functionally impaired hPSC-derived cholangiocytes from cystic fibrosis patients are rescued by CFTR correctors. These findings demonstrate that mature cholangiocytes can be differentiated from hPSCs and used for studies of biliary development and disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
O'Hara, S.P., Tabibian, J.H., Splinter, P.L. & LaRusso, N.F. The dynamic biliary epithelia: molecules, pathways, and disease. J. Hepatol. 58, 575–582 (2013).
Kamath, B.M. et al. Outcomes of liver transplantation for patients with Alagille syndrome: the studies of pediatric liver transplantation experience. Liver Transpl. 18, 940–948 (2012).
Si-Tayeb, K. et al. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology 51, 297–305 (2010).
Touboul, T. et al. Generation of functional hepatocytes from human embryonic stem cells under chemically defined conditions that recapitulate liver development. Hepatology 51, 1754–1765 (2010).
Takayama, K. et al. Generation of metabolically functioning hepatocytes from human pluripotent stem cells by FOXA2 and HNF1alpha transduction. J. Hepatol. 57, 628–636 (2012).
Ogawa, S. et al. Three-dimensional culture and cAMP signaling promote the maturation of human pluripotent stem cell-derived hepatocytes. Development 140, 3285–3296 (2013).
Dianat, N. et al. Generation of functional cholangiocyte-like cells from human pluripotent stem cells and HepaRG cells. Hepatology 60, 700–714 (2014).
Si-Tayeb, K., Lemaigre, F.P. & Duncan, S.A. Organogenesis and development of the liver. Dev. Cell 18, 175–189 (2010).
Zong, Y. et al. Notch signaling controls liver development by regulating biliary differentiation. Development 136, 1727–1739 (2009).
Hofmann, J.J. et al. Jagged1 in the portal vein mesenchyme regulates intrahepatic bile duct development: insights into Alagille syndrome. Development 137, 4061–4072 (2010).
Kodama, Y., Hijikata, M., Kageyama, R., Shimotohno, K. & Chiba, T. The role of notch signaling in the development of intrahepatic bile ducts. Gastroenterology 127, 1775–1786 (2004).
Geisler, F. et al. Liver-specific inactivation of Notch2, but not Notch1, compromises intrahepatic bile duct development in mice. Hepatology 48, 607–616 (2008).
Li, L. et al. Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nat. Genet. 16, 243–251 (1997).
McDaniell, R. et al. NOTCH2 mutations cause Alagille syndrome, a heterogeneous disorder of the notch signaling pathway. Am. J. Hum. Genet. 79, 169–173 (2006).
Kamath, B.M. et al. NOTCH2 mutations in Alagille syndrome. J. Med. Genet. 49, 138–144 (2012).
Gouon-Evans, V. et al. BMP-4 is required for hepatic specification of mouse embryonic stem cell-derived definitive endoderm. Nat. Biotechnol. 24, 1402–1411 (2006).
Zaret, K.S. & Grompe, M. Generation and regeneration of cells of the liver and pancreas. Science 322, 1490–1494 (2008).
Suzuki, A., Sekiya, S., Buscher, D., Izpisua Belmonte, J.C. & Taniguchi, H. Tbx3 controls the fate of hepatic progenitor cells in liver development by suppressing p19ARF expression. Development 135, 1589–1595 (2008).
Lüdtke, T.H., Christoffels, V.M., Petry, M. & Kispert, A. Tbx3 promotes liver bud expansion during mouse development by suppression of cholangiocyte differentiation. Hepatology 49, 969–978 (2009).
Raynaud, P., Carpentier, R., Antoniou, A. & Lemaigre, F.P. Biliary differentiation and bile duct morphogenesis in development and disease. Int. J. Biochem. Cell Biol. 43, 245–256 (2011).
Schmitt, T.M. & Zuniga-Pflucker, J.C. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity 17, 749–756 (2002).
Lehar, S.M., Dooley, J., Farr, A.G. & Bevan, M.J. Notch ligands Delta 1 and Jagged1 transmit distinct signals to T-cell precursors. Blood 105, 1440–1447 (2005).
Fernández-Sánchez, V. et al. In vitro effects of stromal cells expressing different levels of Jagged-1 and Delta-1 on the growth of primitive and intermediate CD34(+) cell subsets from human cord blood. Blood Cells Mol. Dis. 47, 205–213 (2011).
Takayama, K. et al. CCAAT/enhancer binding protein-mediated regulation of TGFbeta receptor 2 expression determines the hepatoblast fate decision. Development 141, 91–100 (2014).
Dorrell, C. et al. Transcriptomes of the major human pancreatic cell types. Diabetologia 54, 2832–2844 (2011).
Irion, S. et al. Identification and targeting of the ROSA26 locus in human embryonic stem cells. Nat. Biotechnol. 25, 1477–1482 (2007).
Dekkers, J.F. et al. A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nat. Med. 19, 939–945 (2013).
Molinski, S. et al. Functional rescue of F508del-CFTR using small molecule correctors. Front. Pharmacol. 3, 160 (2012).
Eckford, P.D., Li, C., Ramjeesingh, M. & Bear, C.E. Cystic fibrosis transmembrane conductance regulator (CFTR) potentiator VX-770 (ivacaftor) opens the defective channel gate of mutant CFTR in a phosphorylation-dependent but ATP-independent manner. J. Biol. Chem. 287, 36639–36649 (2012).
Okiyoneda, T. et al. Mechanism-based corrector combination restores DeltaF508-CFTR folding and function. Nat. Chem. Biol. 9, 444–454 (2013).
Tanimizu, N. & Miyajima, A. Notch signaling controls hepatoblast differentiation by altering the expression of liver-enriched transcription factors. J. Cell Sci. 117, 3165–3174 (2004).
Tanimizu, N., Saito, H., Mostov, K. & Miyajima, A. Long-term culture of hepatic progenitors derived from mouse Dlk+ hepatoblasts. J. Cell Sci. 117, 6425–6434 (2004).
Clotman, F. et al. Control of liver cell fate decision by a gradient of TGF beta signaling modulated by Onecut transcription factors. Genes Dev. 19, 1849–1854 (2005).
Turner, R. et al. Human hepatic stem cell and maturational liver lineage biology. Hepatology 53, 1035–1045 (2011).
Cardinale, V. et al. Multipotent stem/progenitor cells in human biliary tree give rise to hepatocytes, cholangiocytes, and pancreatic islets. Hepatology 54, 2159–2172 (2011).
Leeuwen, L., Fitzgerald, D.A. & Gaskin, K.J. Liver disease in cystic fibrosis. Paediatr. Respir. Rev. 15, 69–74 (2014).
Van Goor, F. et al. Correction of the F508del-CFTR protein processing defect in vitro by the investigational drug VX-809. Proc. Natl. Acad. Sci. USA 108, 18843–18848 (2011).
Zhao, D. et al. Derivation and characterization of hepatic progenitor cells from human embryonic stem cells. PLoS ONE 4, e6468 (2009).
Yanagida, A., Ito, K., Chikada, H., Nakauchi, H. & Kamiya, A. An in vitro expansion system for generation of human iPS cell-derived hepatic progenitor-like cells exhibiting a bipotent differentiation potential. PLoS ONE 8, e67541 (2013).
Kennedy, M., D'Souza, S.L., Lynch-Kattman, M., Schwantz, S. & Keller, G. Development of the hemangioblast defines the onset of hematopoiesis in human ES cell differentiation cultures. Blood 109, 2679–2687 (2007).
Nostro, M.C., Cheng, X., Keller, G.M. & Gadue, P. Wnt, activin, and BMP signaling regulate distinct stages in the developmental pathway from embryonic stem cells to blood. Cell Stem Cell 2, 60–71 (2008).
Nostro, M.C. et al. Stage-specific signaling through TGFbeta family members and WNT regulates patterning and pancreatic specification of human pluripotent stem cells. Development 138, 861–871 (2011).
Wong, A.P. et al. Directed differentiation of human pluripotent stem cells into mature airway epithelia expressing functional CFTR protein. Nat. Biotechnol. 30, 876–882 (2012).
Hotta, A. et al. EOS lentiviral vector selection system for human induced pluripotent stem cells. Nat. Protoc. 4, 1828–1844 (2009).
Acknowledgements
We would like to thank J. Rossant, J. Ellis and A.P. Wong (Hospital for Sick Children, Toronto, ON, Canada) for providing CF patient iPSCs (C1 and GM00997). We thank members of the G.K. and A.G. laboratories for discussion and feedback on the manuscript. We would like to thank F. Xu (Advanced Optical Microscopy Facility, University Health Network) for technical assistance with the time-lapse video. In addition, we would also like to thank O. Adeyi and the members of Department Pathology, University Health Network for technical assistance with immunohistochemistry. H9 hESC was obtained from The Wicell Research Institute (Madison, WI, USA), and MSC-iPSC1 cells were obtained from G.Q. Daley (Harvard Stem Cell Institute). This work was supported by funding from the McEwen Centre for Regenerative Medicine and the Canadian Institutes of Health Research MOP133620, (G.K.), the University Health Network Multi-Organ Transplant Program Academic Enrichment Fund (sponsored by Astellas Pharma Canada), Alagille Syndrome Alliance, SickKids Research Institute, Rare Disease Foundation and the Childhood Liver Disease Research and Education Network, U01 DK062453 (Sokol) from the National Institute of Diabetes, Digestive and Kidney Diseases (B.M.K. and A.G.) and the Canadian Institutes of Health Research (MOP:97954 and GPG:102171 to C.E.B.).
Author information
Authors and Affiliations
Contributions
S.O., M.O., G.K., B.M.K. and A.G. conceptualized the study. S.O. and M.O. led the experimental design and development of the differentiation protocol with input from all authors. S.O. and M.O. performed the hPSC differentiation and characterization of hPSC-derived cholangiocytes. M.O., C.E.B., S.A. and S.C. developed the cyst swelling assay for hPSC-derived cholangiocytes. B.L. and M.G. developed and used the ductal antibody for isolation of primary cholangiocytes from human liver. All authors analyzed and interpreted the data. M.O., S.O., G.K., B.M.K. and A.G. prepared the manuscript with contributions and critical revision from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–11 (PDF 27615 kb)
Supplementary Tables
Supplementary Tables 1–3 (PDF 2152 kb)
41587_2015_BFnbt3294_MOESM26_ESM.mp4
Forskolin/IBMX plus VX-770 induced swelling of C1-iPSCderived cysts in the presence of the CFTR correctors (MP4 2305 kb)
Source data
Rights and permissions
About this article
Cite this article
Ogawa, M., Ogawa, S., Bear, C. et al. Directed differentiation of cholangiocytes from human pluripotent stem cells. Nat Biotechnol 33, 853–861 (2015). https://doi.org/10.1038/nbt.3294
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.3294
This article is cited by
-
Liver Organoids, Novel and Promising Modalities for Exploring and Repairing Liver Injury
Stem Cell Reviews and Reports (2023)
-
A multimodal iPSC platform for cystic fibrosis drug testing
Nature Communications (2022)
-
Genetics, pathobiology and therapeutic opportunities of polycystic liver disease
Nature Reviews Gastroenterology & Hepatology (2022)
-
Biliary atresia-specific deciduous pulp stem cells feature biliary deficiency
Stem Cell Research & Therapy (2021)
-
DL4-μbeads induce T cell lineage differentiation from stem cells in a stromal cell-free system
Nature Communications (2021)