Abstract
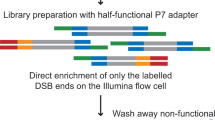
Although great progress has been made in the characterization of the off-target effects of engineered nucleases, sensitive and unbiased genome-wide methods for the detection of off-target cleavage events and potential collateral damage are still lacking. Here we describe a linear amplification–mediated modification of a previously published high-throughput, genome-wide, translocation sequencing (HTGTS) method that robustly detects DNA double-stranded breaks (DSBs) generated by engineered nucleases across the human genome based on their translocation to other endogenous or ectopic DSBs. HTGTS with different Cas9:sgRNA or TALEN nucleases revealed off-target hotspot numbers for given nucleases that ranged from a few or none to dozens or more, and extended the number of known off-targets for certain previously characterized nucleases more than tenfold. We also identified translocations between bona fide nuclease targets on homologous chromosomes, an undesired collateral effect that has not been described previously. Finally, HTGTS confirmed that the Cas9D10A paired nickase approach suppresses off-target cleavage genome-wide.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Sander, J.D. & Joung, J.K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 32, 347–355 (2014).
Kim, H. & Kim, J.S. A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 15, 321–334 (2014).
Yang, H. et al. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154, 1370–1379 (2013).
Yin, H. et al. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat. Biotechnol. 32, 551–553 (2014).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012).
Jinek, M. et al. RNA-programmed genome editing in human cells. eLife 2, e00471 (2013).
Mali, P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013).
Christian, M. et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186, 757–761 (2010).
Miller, J.C. et al. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 29, 143–148 (2011).
Mussolino, C. et al. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 39, 9283–9293 (2011).
Hsu, P.D. et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832 (2013).
Doyle, E.L. et al. TAL Effector-Nucleotide Targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res. 40, W117–122 (2012).
Xiao, A. et al. CasOT: a genome-wide Cas9/sgRNA off-target searching tool. Bioinformatics 30, 1180–1182 (2014).
Fu, Y. et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 31, 822–826 (2013).
Pattanayak, V. et al. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotechnol. 31, 839–843 (2013).
Gabriel, R. et al. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat. Biotechnol. 29, 816–823 (2011).
Petek, L.M., Russell, D.W. & Miller, D.G. Frequent endonuclease cleavage at off-target locations in vivo. Mol. Ther. 18, 983–986 (2010).
Wood, A.J. et al. Targeted genome editing across species using ZFNs and TALENs. Science 333, 307 (2011).
Sung, Y.H. et al. Knockout mice created by TALEN-mediated gene targeting. Nat. Biotechnol. 31, 23–24 (2013).
Mitelman, F., Johansson, B. & Mertens, F. The impact of translocations and gene fusions on cancer causation. Nat. Rev. Cancer 7, 233–245 (2007).
Stephens, P.J. et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature 462, 1005–1010 (2009).
Chiarle, R. et al. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell 147, 107–119 (2011).
Klein, I.A. et al. Translocation-capture sequencing reveals the extent and nature of chromosomal rearrangements in B lymphocytes. Cell 147, 95–106 (2011).
Hu, J., Tepsuporn, S., Meyers, R.M., Gostissa, M. & Alt, F.W. Developmental propagation of V(D)J recombination-associated DNA breaks and translocations in mature B cells via dicentric chromosomes. Proc. Natl. Acad. Sci. USA 111, 10269–10274 (2014).
Barlow, J.H. et al. Identification of early replicating fragile sites that contribute to genome instability. Cell 152, 620–632 (2013).
Zhang, Y. et al. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell 148, 908–921 (2012).
Schmidt, M. et al. High-resolution insertion-site analysis by linear amplification-mediated PCR (LAM-PCR). Nat. Methods 4, 1051–1057 (2007).
Lee, Y.N. et al. A systematic analysis of recombination activity and genotype-phenotype correlation in human recombination-activating gene 1 deficiency. J. Allergy Clin. Immunol. 133, 1099–1108 (2014).
Muñoz, I.G. et al. Molecular basis of engineered meganuclease targeting of the endogenous human RAG1 locus. Nucleic Acids Res. 39, 729–743 (2011).
Krzywinski, M. et al. Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645 (2009).
Hakim, O. et al. DNA damage defines sites of recurrent chromosomal translocations in B lymphocytes. Nature 484, 69–74 (2012).
Alt, F.W., Zhang, Y., Meng, F.L., Guo, C. & Schwer, B. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell 152, 417–429 (2013).
Gostissa, M. et al. IgH class switching exploits a general property of two DNA breaks to be joined in cis over long chromosomal distances. Proc. Natl. Acad. Sci. USA 111, 2644–2649 (2014).
Mali, P. et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 31, 833–838 (2013).
Ran, F.A. et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154, 1380–1389 (2013).
Reyon, D. et al. FLASH assembly of TALENs for high-throughput genome editing. Nat. Biotechnol. 30, 460–465 (2012).
Jiang, W., Bikard, D., Cox, D., Zhang, F. & Marraffini, L.A. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 31, 233–239 (2013).
Asaithamby, A. & Chen, D.J. Cellular responses to DNA double-strand breaks after low-dose gamma-irradiation. Nucleic Acids Res. 37, 3912–3923 (2009).
Franco, S. et al. H2AX prevents DNA breaks from progressing to chromosome breaks and translocations. Mol. Cell 21, 201–214 (2006).
Ramiro, A.R. et al. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature 440, 105–109 (2006).
Hastings, P.J., Lupski, J.R., Rosenberg, S.M. & Ira, G. Mechanisms of change in gene copy number. Nat. Rev. Genet. 10, 551–564 (2009).
Guilinger, J.P. et al. Broad specificity profiling of TALENs results in engineered nucleases with improved DNA-cleavage specificity. Nat. Methods 11, 429–435 (2014).
Zhou, Z.X. et al. Mapping genomic hotspots of DNA damage by a single-strand-DNA-compatible and strand-specific ChIP-seq method. Genome Res. 23, 705–715 (2013).
Faust, G.G. & Hall, I.M. YAHA: fast and flexible long-read alignment with optimal breakpoint detection. Bioinformatics 28, 2417–2424 (2012).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Acknowledgements
This work is supported by the National Institutes of Health Grants P01CA109901 and P01AI076210. R.L.F. was supported by the National Institutes of Health NRSA T32CA070083 and is supported by the National Health Institutes of Health NRSA T32AI007512. J.H. is supported by Robertson Foundation/Cancer Research Institute Irvington Fellowship. F.W.A. is an investigator of the Howard Hughes Medical Institute. A549 cells were provided by David Weinstock (Dana-Farber Cancer Institute).
Author information
Authors and Affiliations
Contributions
R.L.F., J.H. and F.W.A. designed the research; R.L.F., J.H. and E.K. performed the research; R.L.F., J.H., R.M.M., Y.-J.H. and E.K. analyzed the data; R.L.F. and F.W.A. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–16 and Supplementary Tables 1–8 (PDF 9006 kb)
Supplementary Dataset 1
Hotspot listing for HTGTS baits (XLSX 285 kb)
Rights and permissions
About this article
Cite this article
Frock, R., Hu, J., Meyers, R. et al. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat Biotechnol 33, 179–186 (2015). https://doi.org/10.1038/nbt.3101
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.3101
This article is cited by
-
Site-specific transgene integration in chimeric antigen receptor (CAR) T cell therapies
Biomarker Research (2023)
-
CasKAS: direct profiling of genome-wide dCas9 and Cas9 specificity using ssDNA mapping
Genome Biology (2023)
-
Discovering CRISPR–Cas off-target breaks
Nature Methods (2023)
-
Deep sampling of gRNA in the human genome and deep-learning-informed prediction of gRNA activities
Cell Discovery (2023)
-
AAV-mediated delivery of a Sleeping Beauty transposon and an mRNA-encoded transposase for the engineering of therapeutic immune cells
Nature Biomedical Engineering (2023)