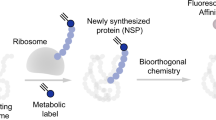
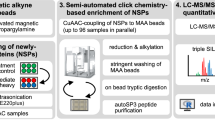
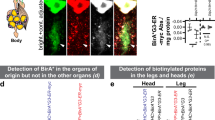
Abstract
Identifying the proteins synthesized at specific times in cells of interest in an animal will facilitate the study of cellular functions and dynamic processes. Here we introduce stochastic orthogonal recoding of translation with chemoselective modification (SORT-M) to address this challenge. SORT-M involves modifying cells to express an orthogonal aminoacyl-tRNA synthetase/tRNA pair to enable the incorporation of chemically modifiable analogs of amino acids at diverse sense codons in cells in rich media. We apply SORT-M to Drosophila melanogaster fed standard food to label and image proteins in specific tissues at precise developmental stages with diverse chemistries, including cyclopropene-tetrazine inverse electron demand Diels-Alder cycloaddition reactions. We also use SORT-M to identify proteins synthesized in germ cells of the fly ovary without dissection. SORT-M will facilitate the definition of proteins synthesized in specific sets of cells to study development, and learning and memory in flies, and may be extended to other animals.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Sopko, R. & Perrimon, N. Receptor tyrosine kinases in Drosophila development. Cold Spring Harb. Perspect. Biol. 10.1101/cshperspect.a009050 (2013).
Keene, A.C. & Sprecher, S.G. Seeing the light: photobehavior in fruit fly larvae. Trends Neurosci. 35, 104–110 (2012).
Tepass, U. The apical polarity protein network in Drosophila epithelial cells: regulation of polarity, junctions, morphogenesis, cell growth, and survival. Annu. Rev. Cell Dev. Biol. 28, 655–685 (2012).
Dubnau, J. Ode to the mushroom bodies. Science 335, 664–665 (2012).
Ngo, J.T. & Tirrell, D.A. Noncanonical amino acids in the interrogation of cellular protein synthesis. Acc. Chem. Res. 44, 677–685 (2011).
Howden, A.J. et al. QuaNCAT: quantitating proteome dynamics in primary cells. Nat. Methods 10, 343–346 (2013).
Ngo, J.T. et al. Cell-selective metabolic labeling of proteins. Nat. Chem. Biol. 5, 715–717 (2009).
Truong, F., Yoo, T.H., Lampo, T.J. & Tirrell, D.A. Two-strain, cell-selective protein labeling in mixed bacterial cultures. J. Am. Chem. Soc. 134, 8551–8556 (2012).
Ngo, J.T., Schuman, E.M. & Tirrell, D.A. Mutant methionyl-tRNA synthetase from bacteria enables site-selective N-terminal labeling of proteins expressed in mammalian cells. Proc. Natl. Acad. Sci. USA 110, 4992–4997 (2013).
Hinz, F.I., Dieterich, D.C., Tirrell, D.A. & Schuman, E.M. Non-canonical amino acid labeling in vivo to visualize and affinity purify newly synthesized proteins in larval zebrafish. ACS Chem. Neurosci. 3, 40–49 (2012).
Link, A.J. et al. Discovery of aminoacyl-tRNA synthetase activity through cell-surface display of noncanonical amino acids. Proc. Natl. Acad. Sci. USA 103, 10180–10185 (2006).
Chin, J.W. Reprogramming the genetic code. Science 336, 428–429 (2012).
Davis, L. & Chin, J.W. Designer proteins: applications of genetic code expansion in cell biology. Nat. Rev. Mol. Cell Biol. 13, 168–182 (2012).
Neumann, H., Peak-Chew, S.Y. & Chin, J.W. Genetically encoding N(epsilon)-acetyllysine in recombinant proteins. Nat. Chem. Biol. 4, 232–234 (2008).
Hancock, S.M., Uprety, R., Deiters, A. & Chin, J.W. Expanding the genetic code of yeast for incorporation of diverse unnatural amino acids via a pyrrolysyl-tRNA synthetase/tRNA pair. J. Am. Chem. Soc. 132, 14819–14824 (2010).
Mukai, T. et al. Adding l–lysine derivatives to the genetic code of mammalian cells with engineered pyrrolysyl-tRNA synthetases. Biochem. Biophys. Res. Commun. 371, 818–822 (2008).
Chen, P.R. et al. A facile system for encoding unnatural amino acids in mammalian cells. Angew. Chem. Int. Ed. 48, 4052–4055 (2009).
Gautier, A. et al. Genetically encoded photocontrol of protein localization in mammalian cells. J. Am. Chem. Soc. 132, 4086–4088 (2010).
Greiss, S. & Chin, J.W. Expanding the genetic code of an animal. J. Am. Chem. Soc. 133, 14196–14199 (2011).
Bianco, A., Townsley, F.M., Greiss, S., Lang, K. & Chin, J.W. Expanding the genetic code of Drosophila melanogaster. Nat. Chem. Biol. 8, 748–750 (2012).
Blackman, M.L., Royzen, M. & Fox, J.M. Tetrazine ligation: fast bioconjugation based on inverse-electron-demand Diels−Alder reactivity. J. Am. Chem. Soc. 130, 13518–13519 (2008).
Yang, J., Šečćkutė, J., Cole, C.M. & Devaraj, N.K. Live-cell imaging of cyclopropene tags with fluorogenic tetrazine cycloadditions. Angew. Chem. Int. Engl.Ed. 51, 7476–7479 (2012).
Cole, C.M., Yang, J., Šečćkutė, J. & Devaraj, N.K. Fluorescent live-cell imaging of metabolically incorporated unnatural cyclopropene-mannosamine derivatives. ChemBioChem 14, 205–208 (2013).
Patterson, D.M., Nazarova, L.A., Xie, B., Kamber, D.N. & Prescher, J.A. Functionalized cyclopropenes as bioorthogonal chemical reporters. J. Am. Chem. Soc. 134, 18638–18643 (2012).
Devaraj, N.K. & Weissleder, R. Biomedical applications of tetrazine cycloadditions. Acc. Chem. Res. 44, 816–827 (2011).
Lang, K. et al. Genetically encoded norbornene directs site-specific cellular protein labelling via a rapid bioorthogonal reaction. Nat. Chem. 4, 298–304 (2012).
Lang, K. et al. Genetic encoding of bicyclononynes and trans-cyclooctenes for site-specific protein labeling in vitro and in live mammalian cells via rapid fluorogenic Diels–Alder reactions. J. Am. Chem. Soc. 134, 10317–10320 (2012).
Borrmann, A. et al. Genetic encoding of a bicyclo[6.1.0]nonyne-charged amino acid enables fast cellular protein imaging by metal-free ligation. ChemBioChem 13, 2094–2099 (2012).
Plass, T. et al. Amino acids for Diels–Alder reactions in living cells. Angew. Chem. Int. Ed. Engl. 51, 4166–4170 (2012).
Seitchik, J.L. et al. Genetically encoded tetrazine amino acid directs rapid site-specific in vivo bioorthogonal ligation with trans-cyclooctenes. J. Am. Chem. Soc. 134, 2898–2901 (2012).
Nguyen, D.P. et al. Genetic encoding and labeling of aliphatic azides and alkynes in recombinant proteins via a pyrrolysyl-tRNA synthetase/tRNACUA pair and click chemistry. J. Am. Chem. Soc. 131, 8720–8721 (2009).
Yu, Z., Pan, Y., Wang, Z., Wang, J. & Lin, Q. Genetically encoded cyclopropene directs rapid, photoclick-chemistry-mediated protein labeling in mammalian cells. Angew. Chem. Int. Ed. Engl. 51, 10600–10604 (2012).
Virdee, S., Ye, Y., Nguyen, D.P., Komander, D. & Chin, J.W. Engineered diubiquitin synthesis reveals Lys29-isopeptide specificity of an OTU deubiquitinase. Nat. Chem. Biol. 6, 750–757 (2010).
Ambrogelly, A. et al. Pyrrolysine is not hardwired for cotranslational insertion at UAG codons. Proc. Natl. Acad. Sci. USA 104, 3141–3146 (2007).
Mosmann, T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods 65, 55–63 (1983).
Brand, A.H. & Perrimon, N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415 (1993).
Hadjieconomou, D. et al. Flybow: genetic multicolor cell labeling for neural circuit analysis in Drosophila melanogaster. Nat. Methods 8, 260–266 (2011).
Wu, J.S. & Luo, L. A protocol for mosaic analysis with a repressible cell marker (MARCM) in Drosophila. Nat. Protoc. 1, 2583–2589 (2006).
Wang, C., Dickinson, L.K. & Lehmann, R. Genetics of nanos localization in Drosophila. Dev. Dyn. 199, 103–115 (1994).
Fox, D.T. & Duronio, R.J. Endoreplication and polyploidy: insights into development and disease. Development 140, 3–12 (2013).
Von Stetina, J.R., Lafever, K.S., Rubin, M. & Drummond-Barbosa, D. A genetic screen for dominant enhancers of the cell-cycle regulator alpha-endosulfine identifies matrimony as a strong functional interactor in Drosophila. G3 (Bethesda) 1, 607–613 (2011).
Feret, R. & Lilley, K. S. Protein profiling using two-dimensional difference gel electrophoresis (2-D Dige). Curr. Protoc. Protein Sci. 75, 22.2 (2014).
Bownes, M. Expression of the genes coding for vitellogenin (yolk protein). Annu. Rev. Entomol. 31, 507–531 (1986).
Graveley, B.R. et al. The developmental transcriptome of Drosophila melanogaster. Nature 471, 473–479 (2011).
Leon, A. & McKearin, D. Identification of TER94, an AAA ATPase protein, as a Bam-dependent component of the Drosophila fusome. Mol. Biol. Cell 10, 3825–3834 (1999).
Liang, L., Diehl-Jones, W. & Lasko, P. Localization of vasa protein to the Drosophila pole plasm is independent of its RNA-binding and helicase activities. Development 120, 1201–1211 (1994).
Suchanek, M., Radzikowska, A. & Thiele, C. Photo-leucine and photo-methionine allow identification of protein-protein interactions in living cells. Nat. Methods 2, 261–268 (2005).
Karp, N.A., Kreil, D.P. & Lilley, K.S. Determining a significant change in protein expression with DeCyder during a pair-wise comparison using two-dimensional difference gel electrophoresis. Proteomics 4, 1421–1432 (2004).
Harper, S., Mozdzanowski, J. & Speicher, D . Two-dimensional gel electrophoresis. Curr. Protoc. Protein Sci. 11, 10.4 (2001).
Acknowledgements
We are exceptionally grateful to M. Skehl and S. Maslen of the LMB Mass Spectrometry service for mass spectrometry, R. Feret of the Cambridge Centre for Proteomics for assistance with DIGE, and S. Bullock (LMB) for advice and comments. We are grateful to the Herchel Smith Fund (S.J.E., administered through Cambridge University Department of Chemistry) the Medical Research Council (U105181009, UD99999908) and the European Research Council for funding.
Author information
Authors and Affiliations
Contributions
T.S.E. developed the chemistry for SORT-M in E. coli with contributions from K.L., A.S. and R.J.E. F.M.T., A.B. and T.S.E. developed SORT-M fly approach. R.J.E. developed SORT-M in mammalian cells with contributions from S.J.E. and L.D. R.P. did initial experiments using 2, with initial input from S.G. K.S.L. provided 2D-DIGE expertise. J.W.C. supervised the project. J.W.C., T.S.E., F.M.T. and A.B. wrote the paper with input from other authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–24, Supplementary Tables 1–3 and Supplementary Note 1 (PDF 4158 kb)
Rights and permissions
About this article
Cite this article
Elliott, T., Townsley, F., Bianco, A. et al. Proteome labeling and protein identification in specific tissues and at specific developmental stages in an animal. Nat Biotechnol 32, 465–472 (2014). https://doi.org/10.1038/nbt.2860
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.2860
This article is cited by
-
Adding α,α-disubstituted and β-linked monomers to the genetic code of an organism
Nature (2024)
-
Quintuply orthogonal pyrrolysyl-tRNA synthetase/tRNAPyl pairs
Nature Chemistry (2023)
-
Light-activated tetrazines enable precision live-cell bioorthogonal chemistry
Nature Chemistry (2022)
-
Extracellular Matrix by Design: Native Biomaterial Fabrication and Functionalization to Boost Tissue Regeneration
Regenerative Engineering and Translational Medicine (2022)
-
Bioorthogonal chemistry
Nature Reviews Methods Primers (2021)