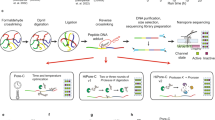
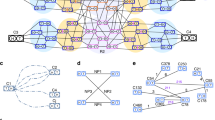
Abstract
Genomes assembled de novo from short reads are highly fragmented relative to the finished chromosomes of Homo sapiens and key model organisms generated by the Human Genome Project. To address this problem, we need scalable, cost-effective methods to obtain assemblies with chromosome-scale contiguity. Here we show that genome-wide chromatin interaction data sets, such as those generated by Hi-C, are a rich source of long-range information for assigning, ordering and orienting genomic sequences to chromosomes, including across centromeres. To exploit this finding, we developed an algorithm that uses Hi-C data for ultra-long-range scaffolding of de novo genome assemblies. We demonstrate the approach by combining shotgun fragment and short jump mate-pair sequences with Hi-C data to generate chromosome-scale de novo assemblies of the human, mouse and Drosophila genomes, achieving—for the human genome—98% accuracy in assigning scaffolds to chromosome groups and 99% accuracy in ordering and orienting scaffolds within chromosome groups. Hi-C data can also be used to validate chromosomal translocations in cancer genomes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature 431, 931–945 (2004).
Shendure, J. & Ji, H. Next-generation DNA sequencing. Nat. Biotechnol. 26, 1135–1145 (2008).
Shendure, J. & Lieberman-Aiden, E. The expanding scope of DNA sequencing. Nat. Biotechnol. 30, 1084–1094 (2012).
Compeau, P., Pevzner, P. & Tesler, G. How to apply de Bruijn graphs to genome assembly. Nat. Biotechnol. 29, 987–991 (2011).
Gnerre, S. et al. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc. Natl. Acad. Sci. USA 108, 1513–1518 (2011).
Li, R. et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 20, 265–272 (2010).
Mouse Genome Sequencing Consortium. Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562 (2002).
Kitzman, J.O. et al. Haplotype-resolved genome sequencing of a Gujarati Indian individual. Nat. Biotechnol. 29, 59–63 (2011).
Zhang, G. et al. The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490, 49–54 (2012).
Schwartz, D.C. et al. Ordered restriction maps of Saccharomyces cerevisiae chromosomes constructed by optical mapping. Science 262, 110–114 (1993).
Zhang, Q. et al. The genome of Prunus mume. Nat. Commun. 3, 1318 (2012).
Dong, Y. et al. Sequencing and automated whole-genome optical mapping of the genome of a domestic goat (Capra hircus). Nat. Biotechnol. 31, 135–141 (2013).
Lam, E. et al. Genome mapping on nanochannel arrays for structural variation analysis and sequence assembly. Nat. Biotechnol. 30, 771–776 (2012).
Baird, N.A. et al. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE 3, e3376 (2008).
Genome 10K Community of Scientists. Genome 10K: a proposal to obtain whole-genome sequence for 10,000 vertebrate species. J. Hered. 100, 659–674 (2009).
Lieberman-Aiden, E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009).
Duan, Z. et al. A three-dimensional model of the yeast genome. Nature 465, 363–367 (2010).
Eisen, M., Spellman, P., Brown, P. & Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95, 14863–14868 (1998).
Dixon, J. et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012).
Yaffe, E. & Tanay, A. Probabilistic modeling of Hi-C contact maps eliminates systematic biases to characterize global chromosomal architecture. Nat. Genet. 43, 1059–1065 (2011).
Mackay, T. et al. The Drosophila melanogaster Genetic Reference Panel. Nature 482, 173–178 (2012).
Sexton, T. et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148, 458–472 (2012).
Landry, J. et al. The genomic and transcriptomic landscape of a HeLa cell line. G3 3, 1213–1224 (2013).
Adey, A. et al. The haplotype-resolved genome and epigenome of the aneuploid HeLa cancer cell line. Nature 500, 207–211 (2013).
Simonis, M. et al. High-resolution identification of balanced and complex chromosomal rearrangements by 4C technology. Nat. Methods 6, 837–842 (2009).
Macville, M. et al. Comprehensive and definitive molecular cytogenetic characterization of HeLa cells by spectral karyotyping. Cancer Res. 59, 141–150 (1999).
Moissiard, G. et al. MORC family ATPases required for heterochromatin condensation and gene silencing. Science 336, 1448–1451 (2012).
Fraley, C. & Raftery, A.E. How many clusters? which clustering method? answers via model-based cluster analysis. Comput. J. 41, 578–588 (1998).
Jung, Y., Park, H., Du, D.Z. & Drake, B. A decision criterion for the optimal number of clusters in hierarchical clustering. J. Glob. Optim. 25, 91–111 (2003).
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010).
Altschul, S.F., Gish, W., Miller, W., Myers, E.W. & Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
van Berkum, N.L. et al. Hi-C: a method to study the three-dimensional architecture of genomes. J. Vis. Exp. 39, e1869 (2010).
Acknowledgements
We thank F. Ay, E. Eichler, J. Felsenstein, P. Green, L. Hillier, M. van Min, W. Noble, R. Waterston and members of the Shendure lab for helpful discussions. Some of the sequencing data used in this research were derived from a HeLa cell line. Henrietta Lacks, and the HeLa cell line that was established from her tumor cells without her knowledge or consent in 1951, have made significant contributions to scientific progress and advances in human health. We are grateful to Henrietta Lacks, now deceased, and to her surviving family members for their contributions to biomedical research. Our work was supported by grant HG006283 from the National Human Genome Research Institute (NHGRI; to J.S.); a graduate research fellowship DGE-0718124 from the National Science Foundation (to A.A. and J.O.K.); and grant T32HG000035 from the NHGRI (to J.N.B.).
Author information
Authors and Affiliations
Contributions
J.N.B., A.A., J.O.K. and J.S. conceived and designed the study. J.N.B. designed and wrote the LACHESIS software. J.N.B. and R.P.P. performed the de novo assemblies. R.Q. conducted the HeLa Hi-C experiments. A.A. analyzed the HeLa Hi-C data. J.N.B., A.A. and J.S. prepared the manuscript, with input from all authors. J.S. supervised the study.
Corresponding authors
Ethics declarations
Competing interests
The authors have fieled a provisional patent application on this method. J.S. is a member of the scientific advisory board or serves as a consultant for Adaptive Biotechnologies, Ariosa Diagnostics, Stratos Genomics, GenePeeks, Gen9, Good Start Genetics, Ingenuity Systems and Rubicon Genomics.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–13 and Supplementary Tables 1–6 (PDF 4599 kb)
Supplementary Data 1
LACHESIS.tar.gz (ZIP 43465 kb)
Supplementary Data 2
README.txt (TXT 13 kb)
Source data
Rights and permissions
About this article
Cite this article
Burton, J., Adey, A., Patwardhan, R. et al. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat Biotechnol 31, 1119–1125 (2013). https://doi.org/10.1038/nbt.2727
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.2727
This article is cited by
-
Chromosome-level genome assembly and annotation of the yellow grouper, Epinephelus awoara
Scientific Data (2024)
-
Chromosome-level genome assembly and annotation of the Yunling cattle with PacBio and Hi-C sequencing data
Scientific Data (2024)
-
Chromosome-level genome assembly of Platycarya strobilacea
Scientific Data (2024)
-
Chromosome-level genome assembly of the diploid oat species Avena longiglumis
Scientific Data (2024)
-
Next-Generation Sequencing in Medicinal Plants: Recent Progress, Opportunities, and Challenges
Journal of Plant Growth Regulation (2024)