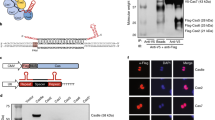
Abstract
Transcription activator–like (TAL) effector nucleases (TALENs) can be readily engineered to bind specific genomic loci, enabling the introduction of precise genetic modifications such as gene knockouts and additions. Here we present a genome-scale collection of TALENs for efficient and scalable gene targeting in human cells. We chose target sites that did not have highly similar sequences elsewhere in the genome to avoid off-target mutations and assembled TALEN plasmids for 18,740 protein-coding genes using a high-throughput Golden-Gate cloning system. A pilot test involving 124 genes showed that all TALENs were active and disrupted their target genes at high frequencies, although two of these TALENs became active only after their target sites were partially demethylated using an inhibitor of DNA methyltransferase. We used our TALEN library to generate single- and double-gene-knockout cells in which NF-κB signaling pathways were disrupted. Compared with cells treated with short interfering RNAs, these cells showed unambiguous suppression of signal transduction.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Venter, J.C. et al. The sequence of the human genome. Science 291, 1304–1351 (2001).
Lander, E.S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Krueger, U. et al. Insights into effective RNAi gained from large-scale siRNA validation screening. Oligonucleotides 17, 237–250 (2007).
Jackson, A.L. et al. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 21, 635–637 (2003).
Birmingham, A. et al. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat. Methods 3, 199–204 (2006).
Khan, A.A. et al. Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat. Biotechnol. 27, 549–555 (2009).
Sledz, C.A., Holko, M., de Veer, M.J., Silverman, R.H. & Williams, B.R. Activation of the interferon system by short-interfering RNAs. Nat. Cell Biol. 5, 834–839 (2003).
Smithies, O., Gregg, R.G., Boggs, S.S., Koralewski, M.A. & Kucherlapati, R.S. Insertion of DNA sequences into the human chromosomal beta-globin locus by homologous recombination. Nature 317, 230–234 (1985).
Bylund, L., Kytola, S., Lui, W.O., Larsson, C. & Weber, G. Analysis of the cytogenetic stability of the human embryonal kidney cell line 293 by cytogenetic and STR profiling approaches. Cytogenet. Genome Res. 106, 28–32 (2004).
Macville, M. et al. Comprehensive and definitive molecular cytogenetic characterization of HeLa cells by spectral karyotyping. Cancer Res. 59, 141–150 (1999).
Mali, P. et al. RNA-Guided Human Genome Engineering via Cas9. Science doi:10.1126/science.1232033 (3 January 2013).
Urnov, F.D., Rebar, E.J., Holmes, M.C., Zhang, H.S. & Gregory, P.D. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 11, 636–646 (2010).
Bibikova, M., Beumer, K., Trautman, J.K. & Carroll, D. Enhancing gene targeting with designed zinc finger nucleases. Science 300, 764 (2003).
Miller, J.C. et al. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 29, 143–148 (2011).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science doi:10.1126/science.1231143 (3 January 2013).
Hwang, W.Y. et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. advance online publication, doi:10.1038/nbt.2501 (29 January 2013).
Cho, S.W., Kim, S., Kim, J.M. & Kim, J.-S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. advance online publication, doi:10.1038/nbt.2507 (29 January 2013).
Kim, H.J., Lee, H.J., Kim, H., Cho, S.W. & Kim, J.S. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 19, 1279–1288 (2009).
Santiago, Y. et al. Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases. Proc. Natl. Acad. Sci. USA 105, 5809–5814 (2008).
Kim, Y.G., Cha, J. & Chandrasegaran, S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. USA 93, 1156–1160 (1996).
Kim, J.S., Lee, H.J. & Carroll, D. Genome editing with modularly assembled zinc-finger nucleases. Nat. Methods 7, 91 author reply 91–92 (2010).
Gupta, A. et al. An optimized two-finger archive for ZFN-mediated gene targeting. Nat. Methods 9, 588–590 (2012).
Reyon, D. et al. FLASH assembly of TALENs for high-throughput genome editing. Nat. Biotechnol. 30, 460–465 (2012).
Carlson, D.F. et al. Efficient TALEN-mediated gene knockout in livestock. Proc. Natl. Acad. Sci. USA 109, 17382–17387 (2012).
Cade, L. et al. Highly efficient generation of heritable zebrafish gene mutations using homo- and heterodimeric TALENs. Nucleic Acids Res. 40, 8001–8010 (2012).
Kim, H., Um, E., Cho, S.R., Jung, C. & Kim, J.S. Surrogate reporters for enrichment of cells with nuclease-induced mutations. Nat. Methods 8, 941–943 (2011).
Li, T. et al. Modularly assembled designer TAL effector nucleases for targeted gene knockout and gene replacement in eukaryotes. Nucleic Acids Res. 39, 6315–6325 (2011).
Zhang, F. et al. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat. Biotechnol. 29, 149–153 (2011).
Cermak, T. et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 39, e82 (2011).
Morbitzer, R., Elsaesser, J., Hausner, J. & Lahaye, T. Assembly of custom TALE-type DNA binding domains by modular cloning. Nucleic Acids Res. 39, 5790–5799 (2011).
Weber, E., Gruetzner, R., Werner, S., Engler, C. & Marillonnet, S. Assembly of designer TAL effectors by Golden Gate cloning. PLoS ONE 6, e19722 (2011).
Guo, J., Gaj, T. & Barbas, C.F. 3rd Directed evolution of an enhanced and highly efficient FokI cleavage domain for zinc finger nucleases. J. Mol. Biol. 400, 96–107 (2010).
Boch, J. et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326, 1509–1512 (2009).
Moscou, M.J. & Bogdanove, A.J. A simple cipher governs DNA recognition by TAL effectors. Science 326, 1501 (2009).
Seal, R.L., Gordon, S.M., Lush, M.J., Wright, M.W. & Bruford, E.A. genenames.org: the HGNC resources in 2011. Nucleic Acids Res. 39, D514–D519 (2011).
Pruitt, K.D., Tatusova, T., Brown, G.R. & Maglott, D.R. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 40, D130–D135 (2012).
Valton, J. et al. Overcoming transcription activator-like effector (TALE) DNA binding domain sensitivity to cytosine methylation. J. Biol. Chem. 287, 38427–38432 (2012).
Deng, D. et al. Recognition of methylated DNA by TAL effectors. Cell Res. 22, 1502–1504 (2012).
Bultmann, S. et al. Targeted transcriptional activation of silent oct4 pluripotency gene by combining designer TALEs and inhibition of epigenetic modifiers. Nucleic Acids Res. 40, 5368–5377 (2012).
Mussolino, C. et al. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 39, 9283–9293 (2011).
Pattanayak, V., Ramirez, C.L., Joung, J.K. & Liu, D.R. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat. Methods 8, 765–770 (2011).
Gabriel, R. et al. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat. Biotechnol. 29, 816–823 (2011).
Lee, H.J., Kim, E. & Kim, J.S. Targeted chromosomal deletions in human cells using zinc finger nucleases. Genome Res. 20, 81–89 (2010).
Tesson, L. et al. Knockout rats generated by embryo microinjection of TALENs. Nat. Biotechnol. 29, 695–696 (2011).
Huang, P. et al. Heritable gene targeting in zebrafish using customized TALENs. Nat. Biotechnol. 29, 699–700 (2011).
Sander, J.D. et al. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat. Biotechnol. 29, 697–698 (2011).
Li, T., Liu, B., Spalding, M.H., Weeks, D.P. & Yang, B. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat. Biotechnol. 30, 390–392 (2012).
Cornu, T.I. et al. DNA-binding specificity is a major determinant of the activity and toxicity of zinc-finger nucleases. Mol. Ther. 16, 352–358 (2008).
Perkins, N.D. Integrating cell-signalling pathways with NF-kappaB and IKK function. Nat. Rev. Mol. Cell Biol. 8, 49–62 (2007).
Volcic, M. et al. NF-kappaB regulates DNA double-strand break repair in conjunction with BRCA1-CtIP complexes. Nucleic Acids Res. 40, 181–195 (2012).
Gewurz, B.E. et al. Genome-wide siRNA screen for mediators of NF-kappaB activation. Proc. Natl. Acad. Sci. USA 109, 2467–2472 (2012).
Briggs, A.W. et al. Iterative capped assembly: rapid and scalable synthesis of repeat-module DNA such as TAL effectors from individual monomers. Nucleic Acids Res. 40, e117 (2012).
Kim, S., Lee, M.J., Kim, H., Kang, M. & Kim, J.S. Preassembled zinc-finger arrays for rapid construction of ZFNs. Nat. Methods 8, 7 (2011).
Sigoillot, F.D. & King, R.W. Vigilance and validation: Keys to success in RNAi screening. ACS Chem. Biol. 6, 47–60 (2011).
Lin, X. et al. siRNA-mediated off-target gene silencing triggered by a 7 nt complementation. Nucleic Acids Res. 33, 4527–4535 (2005).
Adamson, B., Smogorzewska, A., Sigoillot, F.D., King, R.W. & Elledge, S.J. A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nat. Cell Biol. 14, 318–328 (2012).
Holt, N. et al. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat. Biotechnol. 28, 839–847 (2010).
Acknowledgements
This work was supported by the National Research Foundation of Korea (J.-S.K., 2012-0001225), the Intelligent Synthetic Biology Center of the Global Frontier Project funded by the Ministry of Education, Science and Technology, Korea (D.B., 2011-0031956), Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry and Fisheries (S.K., 311062-04-2-sb1010), and Plant Molecular Breeding Center of Next-Generation BioGreen 21 Program (S.K., PJ009081).
Author information
Authors and Affiliations
Contributions
J.-S.K., S.K. and D.B. supervised the research and wrote the manuscript. All the other authors performed the experiments.
Corresponding authors
Ethics declarations
Competing interests
J.Y.Y., M.S.L., E.M.G., H.J.S. and S.K. are employees of ToolGen.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8, Supplementary Tables 2–6 and Supplementary Methods (PDF 2293 kb)
Supplementary Table 1
Supplementary Table 1 (XLSX 100 kb)
Supplementary Table 7
Supplementary Table 7 (XLSX 1501 kb)
Rights and permissions
About this article
Cite this article
Kim, Y., Kweon, J., Kim, A. et al. A library of TAL effector nucleases spanning the human genome. Nat Biotechnol 31, 251–258 (2013). https://doi.org/10.1038/nbt.2517
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.2517
This article is cited by
-
Precision mitochondrial DNA editing with high-fidelity DddA-derived base editors
Nature Biotechnology (2023)
-
Enhanced mitochondrial DNA editing in mice using nuclear-exported TALE-linked deaminases and nucleases
Genome Biology (2022)
-
Targeted A-to-G base editing of chloroplast DNA in plants
Nature Plants (2022)
-
Base editing in human cells with monomeric DddA-TALE fusion deaminases
Nature Communications (2022)
-
Wildtype heterogeneity contributes to clonal variability in genome edited cells
Scientific Reports (2022)