Abstract
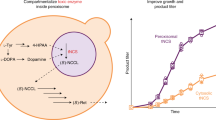
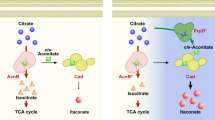
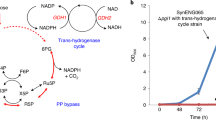
Efforts to improve the production of a compound of interest in Saccharomyces cerevisiae have mainly involved engineering or overexpression of cytoplasmic enzymes. We show that targeting metabolic pathways to mitochondria can increase production compared with overexpression of the enzymes involved in the same pathways in the cytoplasm. Compartmentalization of the Ehrlich pathway into mitochondria increased isobutanol production by 260%, whereas overexpression of the same pathway in the cytoplasm only improved yields by 10%, compared with a strain overproducing enzymes involved in only the first three steps of the biosynthetic pathway. Subcellular fractionation of engineered strains revealed that targeting the enzymes of the Ehrlich pathway to the mitochondria achieves greater local enzyme concentrations. Other benefits of compartmentalization may include increased availability of intermediates, removing the need to transport intermediates out of the mitochondrion and reducing the loss of intermediates to competing pathways.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
03 May 2013
The incorrect version of the supplementary information was posted online and has been replaced as of 3 May 2013.
References
Attardi, G. & Schatz, G. Biogenesis of mitochondria. Annu. Rev. Cell Biol. 4, 289–333 (1988).
Fukuda, H., Casas, A. & Batlle, A. Aminolevulinic acid: from its unique biological function to its star role in photodynamic therapy. Int. J. Biochem. Cell Biol. 37, 272–276 (2005).
Kohlhaw, G.B. Leucine biosynthesis in fungi: entering metabolism through the back door. Microbiol. Mol. Biol. Rev. 67, 1–15 (2003).
Kumar, A. et al. Subcellular localization of the yeast proteome. Genes Dev. 16, 707–719 (2002).
Lange, H., Kispal, G. & Lill, R. Mechanism of iron transport to the site of heme synthesis inside yeast mitochondria. J. Biol. Chem. 274, 18989–18996 (1999).
Marquet, A., Bui, B.T. & Florentin, D. Biosynthesis of biotin and lipoic acid. Vitam. Horm. 61, 51–101 (2001).
Neuburger, M., Rebeille, F., Jourdain, A., Nakamura, S. & Douce, R. Mitochondria are a major site for folate and thymidylate synthesis in plants. J. Biol. Chem. 271, 9466–9472 (1996).
Paltauf, F., Kohlwein, S.D. & Henry, S.A. Regulation and compartmentalization of lipid synthesis in yeast. in The Molecular and Cellular Biology of the Yeast Saccharomyces (eds. Jones, E.W., Pringle, J.R. & Broach, J.R.) 415–500 (Cold Spring Harbor Laboratory Press, 1992).
Pierrel, F. et al. Involvement of mitochondrial ferredoxin and para-aminobenzoic acid in yeast coenzyme Q biosynthesis. Chem. Biol. 17, 449–459 (2010).
Schonauer, M.S., Kastaniotis, A.J., Kursu, V.A., Hiltunen, J.K. & Dieckmann, C.L. Lipoic acid synthesis and attachment in yeast mitochondria. J. Biol. Chem. 284, 23234–23242 (2009).
Shannon, K.W. & Rabinowitz, J.C. Isolation and characterization of the Saccharomyces cerevisiae MIS1 gene encoding mitochondrial C1-tetrahydrofolate synthase. J. Biol. Chem. 263, 7717–7725 (1988).
Sulo, P. & Martin, N.C. Isolation and characterization of LIP5. A lipoate biosynthetic locus of Saccharomyces cerevisiae. J. Biol. Chem. 268, 17634–17639 (1993).
Tran, U.C. & Clarke, C.F. Endogenous synthesis of coenzyme Q in eukaryotes. Mitochondrion 7 (suppl. 7), S62–S71 (2007).
Urban-Grimal, D., Volland, C., Garnier, T., Dehoux, P. & Labbe-Bois, R. The nucleotide sequence of the HEM1 gene and evidence for a precursor form of the mitochondrial 5-aminolevulinate synthase in Saccharomyces cerevisiae. Eur. J. Biochem. 156, 511–519 (1986).
Zhang, S., Sanyal, I., Bulboaca, G.H., Rich, A. & Flint, D.H. The gene for biotin synthase from Saccharomyces cerevisiae: cloning, sequencing, and complementation of Escherichia coli strains lacking biotin synthase. Arch. Biochem. Biophys. 309, 29–35 (1994).
Hiltunen, J.K. et al. Mitochondrial fatty acid synthesis type II: more than just fatty acids. J. Biol. Chem. 284, 9011–9015 (2009).
Stryer, L. Biochemistry, 4th edn (W.H. Freeman and Company, 1995).
Hu, J., Dong, L. & Outten, C.E. The redox environment in the mitochondrial intermembrane space is maintained separately from the cytosol and matrix. J. Biol. Chem. 283, 29126–29134 (2008).
Orij, R., Postmus, J., Ter Beek, A., Brul, S. & Smits, G.J. In vivo measurement of cytosolic and mitochondrial pH using a pH-sensitive GFP derivative in Saccharomyces cerevisiae reveals a relation between intracellular pH and growth. Microbiology 155, 268–278 (2009).
Schnell, N., Krems, B. & Entian, K.D. The PAR1 (YAP1/SNQ3) gene of Saccharomyces cerevisiae, a c-jun homologue, is involved in oxygen metabolism. Curr. Genet. 21, 269–273 (1992).
Muhlenhoff, U. & Lill, R. Biogenesis of iron-sulfur proteins in eukaryotes: a novel task of mitochondria that is inherited from bacteria. Biochim. Biophys. Acta 1459, 370–382 (2000).
Lill, R. & Muhlenhoff, U. Iron-sulfur-protein biogenesis in eukaryotes. Trends Biochem. Sci. 30, 133–141 (2005).
Xu, X.M. & Moller, S.G. Iron-sulfur cluster biogenesis systems and their crosstalk. ChemBioChem 9, 2355–2362 (2008).
Hazelwood, L.A., Daran, J.M., van Maris, A.J., Pronk, J.T. & Dickinson, J.R. The Ehrlich pathway for fusel alcohol production: a century of research on Saccharomyces cerevisiae metabolism. Appl. Environ. Microbiol. 74, 2259–2266 (2008).
Atsumi, S., Hanai, T. & Liao, J.C. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451, 86–89 (2008).
Bastian, S. et al. Engineered ketol-acid reductoisomerase and alcohol dehydrogenase enable anaerobic 2-methylpropan-1-ol production at theoretical yield in Escherichia coli. Metab. Eng. 13, 345–352 (2011).
Higashide, W., Li, Y., Yang, Y. & Liao, J.C. Metabolic engineering of Clostridium cellulolyticum for production of isobutanol from cellulose. Appl. Environ. Microbiol. 77, 2727–2733 (2011).
Jia, X., Li, S., Xie, S. & Wen, J. Engineering a metabolic pathway for isobutanol biosynthesis in Bacillus subtilis. Appl. Biochem. Biotechnol. 168, 1–9 (2012).
Li, S., Wen, J. & Jia, X. Engineering Bacillus subtilis for isobutanol production by heterologous Ehrlich pathway construction and the biosynthetic 2-ketoisovalerate precursor pathway overexpression. Appl. Microbiol. Biotechnol. 91, 577–589 (2011).
Smith, K.M., Cho, K.M. & Liao, J.C. Engineering Corynebacterium glutamicum for isobutanol production. Appl. Microbiol. Biotechnol. 87, 1045–1055 (2010).
Chen, X., Nielsen, K.F., Borodina, I., Kielland-Brandt, M.C. & Karhumaa, K. Increased isobutanol production in Saccharomyces cerevisiae by overexpression of genes in valine metabolism. Biotechnol. Biofuels 4, 21 (2011).
Kondo, T. et al. Genetic engineering to enhance the Ehrlich pathway and alter carbon flux for increased isobutanol production from glucose by Saccharomyces cerevisiae. J. Biotechnol. 159, 32–37 (2012).
Lee, W.H. et al. Isobutanol production in engineered Saccharomyces cerevisiae by overexpression of 2-ketoisovalerate decarboxylase and valine biosynthetic enzymes. Bioprocess Biosyst. Eng. 35, 1467–1475 (2012).
Hong, K.K. & Nielsen, J. Metabolic engineering of Saccharomyces cerevisiae: a key cell factory platform for future biorefineries. Cell Mol. Life Sci. 69, 2671–2690 (2012).
Brat, D., Weber, C., Lorenzen, W., Bode, H.B. & Boles, E. Cytosolic re-localization and optimization of valine synthesis and catabolism enables inseased isobutanol production with the yeast. Saccharomyces cerevisiae. Biotechnol. Biofuels 5, 65 (2012).
Urano, J. et al. US patent. US 2011/0076733 A1 (2011).
Anthony, L.C., Huang, L.L. & Ye, R.W. US patent. US 2010/0129886 A1 (2010).
Buelter, T., Meinhold, P., Smith, C., Aristidou, A., Dundon, C.A. & Urano, J. World patent. WO 2010/075504A2 (2010).
Szczebara, F.M. et al. Total biosynthesis of hydrocortisone from a simple carbon source in yeast. Nat. Biotechnol. 21, 143–149 (2003).
Farhi, M. et al. Harnessing yeast subcellular compartments for the production of plant terpenoids. Metab. Eng. 13, 474–481 (2011).
Maarse, A.C. et al. Subunit IV of yeast cytochrome c oxidase: cloning and nucleotide sequencing of the gene and partial amino acid sequencing of the mature protein. EMBO J. 3, 2831–2837 (1984).
Larroy, C., Pares, X. & Biosca, J.A. Characterization of a Saccharomyces cerevisiae NADP(H)-dependent alcohol dehydrogenase (ADHVII), a member of the cinnamyl alcohol dehydrogenase family. Eur. J. Biochem. 269, 5738–5745 (2002).
Dellomonaco, C., Clomburg, J.M., Miller, E.N. & Gonzalez, R. Engineered reversal of the beta-oxidation cycle for the synthesis of fuels and chemicals. Nature 476, 355–359 (2011).
Feldman, R.M.R. et al. US patent. US 2011/8017375 B2 (2011).
Wu, S. et al. Redirection of cytosolic or plastidic isoprenoid precursors elevates terpene production in plants. Nat. Biotechnol. 24, 1441–1447 (2006).
Christianson, T.W., Sikorski, R.S., Dante, M., Shero, J.H. & Hieter, P. Multifunctional yeast high-copy-number shuttle vectors. Gene 110, 119–122 (1992).
Sleight, S.C., Bartley, B.A., Lieviant, J.A. & Sauro, H.M. In-Fusion BioBrick assembly and re-engineering. Nucleic Acids Res. 38, 2624–2636 (2010).
Blombach, B. & Eikmanns, B.J. Current knowledge on isobutanol production with Escherichia coli, Bacillus subtilis and Corynebacterium glutamicum. Bioeng. Bugs 2, 346–350 (2011).
Gregg, C., Kyryakov, P. & Titorenko, V.I. Purification of mitochondria from yeast cells. J. Vis. Exp. 10.3791/1417 (2009).
Acknowledgements
We thank K.L. Jones Prather and T.D. Fox for helpful discussions, T.J. Helbig for working on GFP subcellular localization experiments, T. DiCesare for preparing figures, S. Lindquist (Whitehead Institute) for strain Y3929, and members of the Stephanopoulos, Fink and Prather laboratories for discussions and advice. J.L.A. is supported by US National Institutes of Health under Ruth L. Kirchstein National Research Service Award 1F32GM098022-01A1. G.R.F. is supported by National Institutes of Health grant GM040266. This work was supported by Shell Global Solutions (US) Inc.
Author information
Authors and Affiliations
Contributions
J.L.A., G.R.F. and G.S. conceived the project, designed the experiments, analyzed the results and wrote the manuscript. J.L.A. designed and made the pJLA vectors, constructed all pathways and strains, and executed all the experiments.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Note, Supplementary Tables 1–5, Supplementary Figures 1–2 (PDF 707 kb)
Rights and permissions
About this article
Cite this article
Avalos, J., Fink, G. & Stephanopoulos, G. Compartmentalization of metabolic pathways in yeast mitochondria improves the production of branched-chain alcohols. Nat Biotechnol 31, 335–341 (2013). https://doi.org/10.1038/nbt.2509
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.2509
This article is cited by
-
Cross-feeding promotes heterogeneity within yeast cell populations
Nature Communications (2024)
-
Microbial host engineering for sustainable isobutanol production from renewable resources
Applied Microbiology and Biotechnology (2024)
-
Research progress of multi-enzyme complexes based on the design of scaffold protein
Bioresources and Bioprocessing (2023)
-
High-level production of nervonic acid in the oleaginous yeast Yarrowia lipolytica by systematic metabolic engineering
Communications Biology (2023)
-
Spatiotemporal control for integrated catalysis
Nature Reviews Methods Primers (2023)