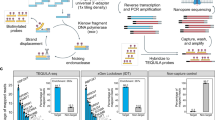
Abstract
Transcriptomic analyses have revealed an unexpected complexity to the human transcriptome, whose breadth and depth exceeds current RNA sequencing capability1,2,3,4. Using tiling arrays to target and sequence select portions of the transcriptome, we identify and characterize unannotated transcripts whose rare or transient expression is below the detection limits of conventional sequencing approaches. We use the unprecedented depth of coverage afforded by this technique to reach the deepest limits of the human transcriptome, exposing widespread, regulated and remarkably complex noncoding transcription in intergenic regions, as well as unannotated exons and splicing patterns in even intensively studied protein-coding loci such as p53 and HOX. The data also show that intermittent sequenced reads observed in conventional RNA sequencing data sets, previously dismissed as noise, are in fact indicative of unassembled rare transcripts. Collectively, these results reveal the range, depth and complexity of a human transcriptome that is far from fully characterized.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
References
Birney, E. et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447, 799–816 (2007).
Carninci, P. et al. The transcriptional landscape of the mammalian genome. Science 309, 1559–1563 (2005).
Katayama, S. et al. Antisense transcription in the mammalian transcriptome. Science 309, 1564–1566 (2005).
van Bakel, H., Nislow, C., Blencowe, B.J. & Hughes, T.R. Response to “the reality of pervasive transcription”. PLoS Biol. 9, e1001102 (2011).
Clark, M.B. et al. The reality of pervasive transcription. PLoS Biol. 9, e1000625 (2011).
van Bakel, H., Nislow, C., Blencowe, B.J. & Hughes, T.R. Most “dark matter” transcripts are associated with known genes. PLoS Biol. 8, e1000371 (2010).
Levin, J.Z. et al. Targeted next-generation sequencing of a cancer transcriptome enhances detection of sequence variants and novel fusion transcripts. Genome Biol. 10, R115 (2009).
Teer, J.K. et al. Systematic comparison of three genomic enrichment methods for massively parallel DNA sequencing. Genome Res. 20, 1420–1431 (2010).
Yehle, C.O. et al. A solution hybridization assay for ribosomal RNA from bacteria using biotinylated DNA probes and enzyme-labeled antibody to DNA:RNA. Mol. Cell. Probes 1, 177–193 (1987).
Crider-Miller, S.J. et al. Novel transcribed sequences within the BWS/WT2 region in 11p15.5: tissue-specific expression correlates with cancer type. Genomics 46, 355–363 (1997).
Rinn, J.L., Bondre, C., Gladstone, H.B., Brown, P.O. & Chang, H.Y. Anatomic demarcation by positional variation in fibroblast gene expression programs. PLoS Genet. 2, e119 (2006).
Trapnell, C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010).
Rinn, J.L. et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129, 1311–1323 (2007).
Li, Y. et al. Resequencing of 200 human exomes identifies an excess of low-frequency non-synonymous coding variants. Nat. Genet. 42, 969–972 (2010).
Ng, S.B. et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature 461, 272–276 (2009).
Kapranov, P. et al. Examples of the complex architecture of the human transcriptome revealed by RACE and high-density tiling arrays. Genome Res. 15, 987–997 (2005).
Pan, Q., Shai, O., Lee, L.J., Frey, B.J. & Blencowe, B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 40, 1413–1415 (2008).
Khoury, M.P. & Bourdon, J.C. The isoforms of the p53 protein. Cold Spring Harb. Perspect. Biol. 2, a000927 (2010).
Olivares-Illana, V. & Fahraeus, R. p53 isoforms gain functions. Oncogene 29, 5113–5119 (2010).
Kloc, M., Foreman, V. & Reddy, S.A. Binary function of mRNA. Biochimie 93, 1955–1961 (2011).
Mercer, T.R., Dinger, M.E. & Mattick, J.S. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 10, 155–159 (2009).
Tsai, M.C. et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science 329, 689–693 (2010).
Hiller, M. & Platzer, M. Widespread and subtle: alternative splicing at short-distance tandem sites. Trends Genet. 24, 246–255 (2008).
Schatz, M.C., Delcher, A.L. & Salzberg, S.L. Assembly of large genomes using second-generation sequencing. Genome Res. 20, 1165–1173 (2010).
Khalil, A.M. et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. USA 106, 11667–11672 (2009).
Trapnell, C., Pachter, L. & Salzberg, S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009).
Carter, M.G. et al. Transcript copy number estimation using a mouse whole-genome oligonucleotide microarray. Genome Biol. 6, R61 (2005).
Hindorff, L.A. et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. USA 106, 9362–9367 (2009).
Islam, S. et al. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res. 21, 1160–1167 (2011).
Hah, N. et al. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell 145, 622–634 (2011).
Morgulis, A., Gertz, E.M., Schaffer, A.A. & Agarwala, R. WindowMasker: window-based masker for sequenced genomes. Bioinformatics 22, 134–141 (2006).
Fu, Y. et al. Repeat subtraction-mediated sequence capture from a complex genome. Plant J. 62, 898–909 (2010).
Quinlan, A.R. & Hall, I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Siepel, A. et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15, 1034–1050 (2005).
Kong, L. et al. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 35, W345 (2007).
Lin, M.F. et al. Revisiting the protein-coding gene catalog of Drosophila melanogaster using 12 fly genomes. Genome Res. 17, 1823–1836 (2007).
Suzek, B.E., Huang, H., McGarvey, P., Mazumder, R. & Wu, C.H. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics 23, 1282–1288 (2007).
Acknowledgements
The authors would like to thank M. Garber for his design of the arrays and constructive contribution to the manuscript; M. Koziol and K. Thomas for preparing cDNA and sequencing libraries; T. Albert, T. Arnold, J. Affourtit, B. Dessany, T. Jarvie, D. Green and T. Millard provided sequencing support. The authors would like to thank the following funding sources: Human Frontiers Science Program (to T.R.M.); Queensland Government Department of Employment, Economic Development and Innovation Smart Futures Fellowship (to M.E.D.); Australian Research Council/University of Queensland co-sponsored Federation Fellowship (FF0561986; to J.S.M.); Australian National Health and Medical Research Council Australia Fellowship (631668; to J.S.M.) and Career Development Award (CDA631542; to M.E.D.); Damon Runyon-Rachleff, Searle, Smith Family Foundation and Richard Merkin Foundation Scholar (to J.L.R.); and US National Institutes of Health (1DP2OD00667-01; to J.L.R. and C.T.).
Author information
Authors and Affiliations
Contributions
T.R.M., J.A.J., J.S.M. and J.L.R. designed the experiments. D.J.G. performed array capture, quality assessments and supported the sequencing teams. J.C. performed RT-PCR. M.E.D., T.R.M. and C.T. performed alignment, transcript assembly and analysis. T.R.M., M.E.D., J.A.J., J.S.M. and J.L.R. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
T.R.M., M.E.D., C.T., J.C., J.S.M. and J.L.R. declare no competing financial interests. D.J.G. and J.A.J are employees of Roche NimbleGen, Inc.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 and Supplementary Results (PDF 1707 kb)
Supplementary Tables 1-4
Summary of library sequencing and alignment employed within study. (XLSX 219 kb)
Supplementary Data
Size and genome corrdinates of probed regions (ZIP 3238 kb)
Rights and permissions
About this article
Cite this article
Mercer, T., Gerhardt, D., Dinger, M. et al. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat Biotechnol 30, 99–104 (2012). https://doi.org/10.1038/nbt.2024
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.2024
This article is cited by
-
The BulkECexplorer compiles endothelial bulk transcriptomes to predict functional versus leaky transcription
Nature Cardiovascular Research (2024)
-
Blocking Abundant RNA Transcripts by High-Affinity Oligonucleotides during Transcriptome Library Preparation
Biological Procedures Online (2023)
-
Evidence for widespread existence of functional novel and non-canonical human transcripts
BMC Biology (2023)
-
Methodological considerations for aqueous environmental RNA collection, preservation, and extraction
Analytical Sciences (2023)
-
Non-coding RNAs in human health and disease: potential function as biomarkers and therapeutic targets
Functional & Integrative Genomics (2023)