Abstract
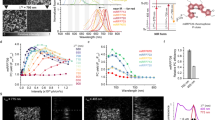
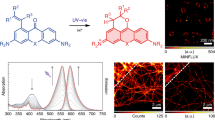
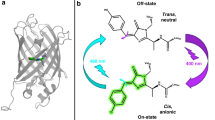
Photoswitchable fluorescent proteins have enabled new approaches for imaging cells, but their utility has been limited either because they cannot be switched repeatedly or because the wavelengths for switching and fluorescence imaging are strictly coupled. We report a bright, monomeric, reversibly photoswitchable variant of GFP, Dreiklang, whose fluorescence excitation spectrum is decoupled from that for optical switching. Reversible on-and-off switching in living cells is accomplished at illumination wavelengths of ∼365 nm and ∼405 nm, respectively, whereas fluorescence is elicited at ∼515 nm. Mass spectrometry and high-resolution crystallographic analysis of the same protein crystal in the photoswitched on- and off-states demonstrate that switching is based on a reversible hydration/dehydration reaction that modifies the chromophore. The switching properties of Dreiklang enable far-field fluorescence nanoscopy in living mammalian cells using both a coordinate-targeted and a stochastic single molecule switching approach.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Tsien, R.Y. The green fluorescent protein. Annu. Rev. Biochem. 67, 509–544 (1998).
Lippincott-Schwartz, J., Altan-Bonnet, N. & Patterson, G.H. Photobleaching and photoactivation: following protein dynamics in living cells. Nat. Cell Biol. S7–S14 (2003).
Lukyanov, K.A., Chudakov, D.M., Lukyanov, S. & Verkhusha, V.V. Innovation: photoactivatable fluorescent proteins. Nat. Rev. Mol. Cell Biol. 6, 885–890 (2005).
Dickson, R.M., Cubitt, A.B., Tsien, R.Y. & Moerner, W.E. On/off blinking and switching behaviour of single molecules of green fluorescent protein. Nature 388, 355–358 (1997).
Habuchi, S. et al. Reversible single-molecule photoswitching in the GFP-like fluorescent protein Dronpa. Proc. Natl. Acad. Sci. USA 102, 9511–9516 (2005).
Hell, S.W., Jakobs, S. & Kastrup, L. Imaging and writing at the nanoscale with focused visible light through saturable optical transitions. Appl. Phys. A Mater. Sci. Process. 77, 859–860 (2003).
Hell, S.W. Toward fluorescence nanoscopy. Nat. Biotechnol. 21, 1347–1355 (2003).
Hofmann, M., Eggeling, C., Jakobs, S. & Hell, S.W. Breaking the diffraction barrier in fluorescence microscopy at low light intensities by using reversibly photoswitchable proteins. Proc. Natl. Acad. Sci. USA 102, 17565–17569 (2005).
Betzig, E. et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–1645 (2006).
Hess, S.T., Girirajan, T.P. & Mason, M.D. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys. J. 91, 4258–4272 (2006).
Patterson, G.H. & Lippincott-Schwartz, J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science 297, 1873–1877 (2002).
Chudakov, D.M. et al. Photoswitchable cyan fluorescent protein for protein tracking. Nat. Biotechnol. 22, 1435–1439 (2004).
Subach, F.V. et al. Photoactivatable mCherry for high-resolution two-color fluorescence microscopy. Nat. Methods 6, 153–159 (2009).
Lukyanov, K.A. et al. Natural animal coloration can be determined by a nonfluorescent green fluorescent protein homolog. J. Biol. Chem. 275, 25879–25882 (2000).
Cinelli, R.A.G. et al. Green fluorescent proteins as optically controllable elements in bioelectronics. Appl. Phys. Lett. 79, 3353–3355 (2001).
Ando, R., Mizuno, H. & Miyawaki, A. Regulated fast nucleocytoplasmic shuttling observed by reversible protein highlighting. Science 306, 1370–1373 (2004).
Henderson, J.N., Ai, H.W., Campbell, R.E. & Remington, S.J. Structural basis for reversible photobleaching of a green fluorescent protein homologue. Proc. Natl. Acad. Sci. USA 104, 6672–6677 (2007).
Stiel, A.C. et al. 1.8 Å bright-state structure of the reversibly switchable fluorescent protein Dronpa guides the generation of fast switching variants. Biochem. J. 402, 35–42 (2007).
Andresen, M. et al. Photoswitchable fluorescent proteins enable monochromatic multilabel imaging and dual color fluorescence nanoscopy. Nat. Biotechnol. 26, 1035–1040 (2008).
Stiel, A.C. et al. Generation of monomeric reversibly switchable red fluorescent proteins for far-field fluorescence nanoscopy. Biophys. J. 95, 2989–2997 (2008).
Adam, V. et al. Structural characterization of IrisFP, an optical highlighter undergoing multiple photo-induced transformations. Proc. Natl. Acad. Sci. USA 105, 18343–18348 (2008).
Subach, F.V. et al. Red fluorescent protein with reversibly photoswitchable absorbance for photochromic FRET. Chem. Biol. 17, 745–755 (2010).
Sinnecker, D., Voigt, P., Hellwig, N. & Schaefer, M. Reversible photobleaching of enhanced green fluorescent proteins. Biochemistry 44, 7085–7094 (2005).
Shaner, N.C. et al. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat. Methods 5, 545–551 (2008).
Ormö, M. et al. Crystal structure of the Aequorea victoria green fluorescent protein. Science 273, 1392–1395 (1996).
Griesbeck, O., Baird, G.S., Campbell, R.E., Zacharias, D.A. & Tsien, R.Y. Reducing the environmental sensitivity of yellow fluorescent protein – Mechanism and applications. J. Biol. Chem. 276, 29188–29194 (2001).
Siegbahn, P.E.M., Wirstam, M. & Zimmer, M. Theoretical study of the mechanism of peptide ring formation in green fluorescent protein. Int. J. Quantum Chem. 81, 169–186 (2001).
Rosenow, M.A., Huffman, H.A., Phail, M.E. & Wachter, R.M. The crystal structure of the Y66L variant of green fluorescent protein supports a cyclization-oxidation-dehydration mechanism for chromophore maturation. Biochemistry 43, 4464–4472 (2004).
Barondeau, D.P., Kassmann, C.J., Tainer, J.A. & Getzoff, E.D. Understanding GFP chromophore biosynthesis: controlling backbone cyclization and modifying post-translational chemistry. Biochemistry 44, 1960–1970 (2005).
Irie, M., Fukaminato, T., Sasaki, T., Tamai, N. & Kawai, T. Organic chemistry: a digital fluorescent molecular photoswitch. Nature 420, 759–760 (2002).
Sauer, M. Reversible molecular photoswitches: a key technology for nanoscience and fluorescence imaging. Proc. Natl. Acad. Sci. USA 102, 9433–9434 (2005).
Lippincott-Schwartz, J., Snapp, E. & Kenworthy, A. Studying protein dynamics in living cells. Nat. Rev. Mol. Cell Biol. 2, 444–456 (2001).
Phair, R.D. & Misteli, T. Kinetic modelling approaches to in vivo imaging. Nat. Rev. Mol. Cell Biol. 2, 898–907 (2001).
Sprague, B.L. & McNally, J.G. FRAP analysis of binding: proper and fitting. Trends Cell Biol. 15, 84–91 (2005).
Chudakov, D.M., Chepurnykh, T.V., Belousov, V.V., Lukyanov, S. & Lukyanov, K.A. Fast and precise protein tracking using repeated reversible photoactivation. Traffic 7, 1304–1310 (2006).
Hell, S.W. Microscopy and its focal switch. Nat. Methods 6, 24–32 (2009).
Rust, M., Bates, M. & Zhuang, X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–796 (2006).
Lamesch, P. et al. hORFeome v3.1: a resource of human open reading frames representing over 10,000 human genes. Genomics 89, 307–315 (2007).
Testa, I. et al. Nanoscale separation of molecular species based on their rotational mobility. Opt. Express 16, 21093–21104 (2008).
Patterson, G.H., Knobel, S.M., Sharif, W.D., Kain, S.R. & Piston, D.W. Use of the green fluorescent protein and its mutants in quantitative fluorescence microscopy. Biophys. J. 73, 2782–2790 (1997).
Bossi, M. et al. Multicolor far-field fluorescence nanoscopy through isolated detection of distinct molecular species. Nano Lett. 8, 2463–2468 (2008).
Richter, F.M., Sander, B., Golas, M.M., Stark, H. & Urlaub, H. Merging molecular electron microscopy and mass spectrometry by carbon film-assisted endoproteinase digestion. Mol. Cell. Proteomics 9, 1729–1741 (2010).
Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 26, 795–800 (1993).
McCoy, A.J. et al. Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Adams, P.D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Schuttelkopf, A.W. & van Aalten, D.M. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 60, 1355–1363 (2004).
Moriarty, N.W., Grosse-Kunstleve, R.W. & Adams, P.D. electronic ligand builder and optimization workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Crystallogr. D Biol. Crystallogr. 65, 1074–1080 (2009).
Acknowledgements
We acknowledge access to beamline BL14.2 of the BESSY II storage ring (Berlin) through the Joint Berlin MX-Laboratory sponsored by the Helmholtz Zentrum Berlin für Materialien und Energie, the Freie Universität Berlin, the Humboldt-Universität zu Berlin, the Max-Delbrück Centrum and the Leibniz-Institut für Molekulare Pharmakologie. We thank V. Belov for insightful discussions and F. Lavoie-Cardinal for help with the set-up for targeted switching. We acknowledge A. Schönle for providing the software ImSpector. We also thank T. Gilat, S. Löbermann, R. Pick and E. Rothermel for excellent technical assistance, H.-H. Hsiao for help in ESI-MS and H. Schill and J. Jethwa for carefully reading the manuscript. We acknowledge R.Y. Tsien for sharing the plasmid pRSET-Citrine. This work was supported by the Deutsche Forschungsgemeinschaft through the Gottfried Wilhelm Leibniz Prize (to S.W.H.) and through the DFG-Research Center for Molecular Physiology of the Brain (to S.J.).
Author information
Authors and Affiliations
Contributions
G.W., M.A. and I.T. contributed equally to this work. C.E., S.W.H. and S.J. conceived the project. T.B., A.C.S., G.W., M.A., I.T., T.G., M.L. and U.P. performed all experiments. I.T. recorded the super-resolution images. Data analysis was done by T.B., A.C.S., G.W., M.A., I.T., T.G., M.L., H.U., C.E., M.C.W., S.W.H. and S.J. The manuscript was written by S.W.H. and S.J. All authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
A patent application concerning the protein Dreiklang has been filed.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1,2 and Supplementary and Figures 1–17 (PDF 1847 kb)
Supplementary Movie 1
Animated sequence of 33 individual images as shown in Figure 3a. (MPG 232 kb)
Supplementary Movie 2
Animated sequence of 100 consecutive switching cycles of vimentin-Dreiklang in PtK2 cells, as shown in Supplementary Figure 13. (AVI 1396 kb)
Rights and permissions
About this article
Cite this article
Brakemann, T., Stiel, A., Weber, G. et al. A reversibly photoswitchable GFP-like protein with fluorescence excitation decoupled from switching. Nat Biotechnol 29, 942–947 (2011). https://doi.org/10.1038/nbt.1952
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.1952
This article is cited by
-
Near-infrared PAINT localization microscopy via chromophore replenishment of phytochrome-derived fluorescent tag
Communications Biology (2024)
-
Indefinite and bidirectional near-infrared nanocrystal photoswitching
Nature (2023)
-
Multiphoton single-molecule localization by sequential excitation with light minima
Light: Science & Applications (2022)
-
First biphotochromic fluorescent protein moxSAASoti stabilized for oxidizing environment
Scientific Reports (2022)
-
Extension of the short wavelength side of fluorescent proteins using hydrated chromophores, and its application
Communications Biology (2022)