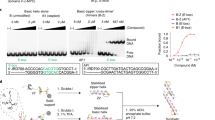
Abstract
The ability to direct functional proteins to specific DNA sequences is a long-sought goal in the study and engineering of biological processes. Transcription activator–like effectors (TALEs) from Xanthomonas sp. are site-specific DNA-binding proteins that can be readily designed to target new sequences. Because TALEs contain a large number of repeat domains, it can be difficult to synthesize new variants. Here we describe a method that overcomes this problem. We leverage codon degeneracy and type IIs restriction enzymes to generate orthogonal ligation linkers between individual repeat monomers, thus allowing full-length, customized, repeat domains to be constructed by hierarchical ligation. We synthesized 17 TALEs that are customized to recognize specific DNA-binding sites, and demonstrate that they can specifically modulate transcription of endogenous genes (SOX2 and KLF4) in human cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Carr, P.A. & Church, G.M. Genome engineering. Nat. Biotechnol. 27, 1151–1162 (2009).
Meister, G.E., Chandrasegaran, S. & Ostermeier, M. Heterodimeric DNA methyltransferases as a platform for creating designer zinc finger methyltransferases for targeted DNA methylation in cells. Nucleic Acids Res. 38, 1749–1759 (2010).
Gonzalez, B. et al. Modular system for the construction of zinc-finger libraries and proteins. Nat. Protoc. 5, 791–810 (2010).
Maeder, M.L., Thibodeau-Beganny, S., Sander, J.D., Voytas, D.F. & Joung, J.K. Oligomerized pool engineering (OPEN): an 'open-source' protocol for making customized zinc-finger arrays. Nat. Protoc. 4, 1471–1501 (2009).
Blancafort, P., Magnenat, L. & Barbas, C.F., III . Scanning the human genome with combinatorial transcription factor libraries. Nat. Biotechnol. 21, 269–274 (2003).
Rosen, L.E. et al. Homing endonuclease I-CreI derivatives with novel DNA target specificities. Nucleic Acids Res. 34, 4791–4800 (2006).
Grizot, S. et al. Efficient targeting of a SCID gene by an engineered single-chain homing endonuclease. Nucleic Acids Res. 37, 5405–5419 (2009).
Boch, J. et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326, 1509–1512 (2009).
Boch, J. & Bonas, U. Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu. Rev. Phytopathol. 48, 419–436 (2010).
Moscou, M.J. & Bogdanove, A.J. A simple cipher governs DNA recognition by TAL effectors. Science 326, 1501 (2009).
Bogdanove, A.J., Schornack, S. & Lahaye, T. TAL effectors: finding plant genes for disease and defense. Curr. Opin. Plant Biol. 13, 394–401 (2010).
Morbitzer, R., Romer, P., Boch, J. & Lahaye, T. Regulation of selected genome loci using de novo-engineered transcription activator-like effector (TALE)-type transcription factors. Proc. Natl. Acad. Sci. USA 107, 21617–21622 (2010).
Kay, S., Hahn, S., Marois, E., Wieduwild, R. & Bonas, U. Detailed analysis of the DNA recognition motifs of the Xanthomonas type III effectors AvrBs3 and AvrBs3Deltarep16. Plant J. 59, 859–871 (2009).
Romer, P. et al. Recognition of AvrBs3-like proteins is mediated by specific binding to promoters of matching pepper Bs3 alleles. Plant Physiol. 150, 1697–1712 (2009).
Kay, S., Hahn, S., Marois, E., Hause, G. & Bonas, U. A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science 318, 648–651 (2007).
Romer, P. et al. Plant pathogen recognition mediated by promoter activation of the pepper Bs3 resistance gene. Science 318, 645–648 (2007).
Scholze, H. & Boch, J. TAL effector-DNA specificity. Virulence 1, 428–432 (2010).
Christian, M. et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186, 757–761 (2010).
Miller, J.C. et al. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. published online, doi:10.1038/nbt.1755 (22 December 2010).
Engler, C., Gruetzner, R., Kandzia, R. & Marillonnet, S. Golden gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS ONE 4, e5553 (2009).
Engler, C., Kandzia, R. & Marillonnet, S. A one pot, one step, precision cloning method with high throughput capability. PLoS ONE 3, e3647 (2008).
Beerli, R.R., Segal, D.J., Dreier, B. & Barbas, C.F., III . Toward controlling gene expression at will: specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc. Natl. Acad. Sci. USA 95, 14628–14633 (1998).
Xu, N., Papagiannakopoulos, T., Pan, G., Thomson, J.A. & Kosik, K.S. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell 137, 647–658 (2009).
Wei, Z. et al. Klf4 interacts directly with Oct4 and Sox2 to promote reprogramming. Stem Cells 27, 2969–2978 (2009).
Lee, H.J., Kim, E. & Kim, J.S. Targeted chromosomal deletions in human cells using zinc finger nucleases. Genome Res. 20, 81–89 (2010).
Li, T. et al. TAL nucleases (TALNs): hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain. Nucleic Acids Res. 39, 359–372 (2010).
Gordley, R.M., Gersbach, C.A. & Barbas, C.F., III . Synthesis of programmable integrases. Proc. Natl. Acad. Sci. USA 106, 5053–5058 (2009).
Zhang, F. et al. Multimodal fast optical interrogation of neural circuitry. Nature 446, 633–639 (2007).
Acknowledgements
This work was supported by the Harvard Society of Fellows (F.Z.), National Human Genome Research Institute Center for Excellence in Genomics Science (P50 HG003170, G.M.C.), Department of Energy Genomes to Life (DE-FG02-02ER63445, G.M.C.), Defense Advanced Research Projects Agency (W911NF-08-1-0254, G.M.C.), the Wyss Institute for Biologically Inspired Engineering (G.M.C.) and National Institutes of Health Transformative R01 (R01 NS073124-01, F.Z. and P.A.). S.L. was partially supported by a predoctoral fellowship from the European School of Molecular Medicine (S.E.M.M.). We thank the entire Church and Arlotta laboratories for discussion and support.
Author information
Authors and Affiliations
Contributions
F.Z. and L.C. conceived the study. F.Z., L.C., S.L. and S.K. designed, performed and analyzed all experiments. P.A. supervised the work of S.L. and G.M.C. supervised the work of F.Z., L.C. and S.K. G.M.C., P.A. and F.Z. provided support for this study. F.Z., L.C. and P.A. wrote the manuscript with support from all authors. G.M.C and P.A. equally contributed to this work.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–3, Supplementary Figs. 1–3, Supplementary Methods and Supplementary Sequences (PDF 804 kb)
Rights and permissions
About this article
Cite this article
Zhang, F., Cong, L., Lodato, S. et al. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat Biotechnol 29, 149–153 (2011). https://doi.org/10.1038/nbt.1775
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.1775
This article is cited by
-
Live-cell imaging of chromatin contacts opens a new window into chromatin dynamics
Epigenetics & Chromatin (2023)
-
Toward improved terpenoids biosynthesis: strategies to enhance the capabilities of cell factories
Bioresources and Bioprocessing (2022)
-
PERSIST platform provides programmable RNA regulation using CRISPR endoRNases
Nature Communications (2022)