Abstract
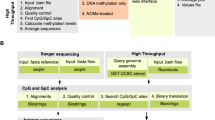
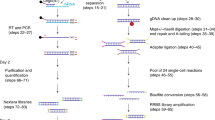
Analysis of DNA methylation patterns relies increasingly on sequencing-based profiling methods. The four most frequently used sequencing-based technologies are the bisulfite-based methods MethylC-seq and reduced representation bisulfite sequencing (RRBS), and the enrichment-based techniques methylated DNA immunoprecipitation sequencing (MeDIP-seq) and methylated DNA binding domain sequencing (MBD-seq). We applied all four methods to biological replicates of human embryonic stem cells to assess their genome-wide CpG coverage, resolution, cost, concordance and the influence of CpG density and genomic context. The methylation levels assessed by the two bisulfite methods were concordant (their difference did not exceed a given threshold) for 82% for CpGs and 99% of the non-CpG cytosines. Using binary methylation calls, the two enrichment methods were 99% concordant and regions assessed by all four methods were 97% concordant. We combined MeDIP-seq with methylation-sensitive restriction enzyme (MRE-seq) sequencing for comprehensive methylome coverage at lower cost. This, along with RNA-seq and ChIP-seq of the ES cells enabled us to detect regions with allele-specific epigenetic states, identifying most known imprinted regions and new loci with monoallelic epigenetic marks and monoallelic expression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Robertson, K.D. DNA methylation and human disease. Nat. Rev. Genet. 6, 597–610 (2005).
Bird, A. DNA methylation patterns and epigenetic memory. Genes Dev. 16, 6–21 (2002).
Feinberg, A.P. & Vogelstein, B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature 301, 89–92 (1983).
Gama-Sosa, M.A. et al. Tissue-specific differences in DNA methylation in various mammals. Biochim. Biophys. Acta 740, 212–219 (1983).
Tahiliani, M. et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935 (2009).
Kriaucionis, S. & Heintz, N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324, 929–930 (2009).
Ito, S. et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466, 1129–1133 (2010).
Lister, R. et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462, 315–322 (2009).
Meissner, A. et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454, 766–770 (2008).
Jacinto, F.V., Ballestar, E. & Esteller, M. Methyl-DNA immunoprecipitation (MeDIP): hunting down the DNA methylome. Biotechniques 44, 35–43 (2008).
Down, T.A. et al. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat. Biotechnol. 26, 779–785 (2008).
Serre, D., Lee, B.H. & Ting, A.H. MBD-isolated Genome Sequencing provides a high-throughput and comprehensive survey of DNA methylation in the human genome. Nucleic Acids Res. 38, 391–399 (2010).
Maunakea, A.K. et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 466, 253–257 (2010).
Ball, M.P. et al. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat. Biotechnol. 27, 361–368 (2009).
Cokus, S.J. et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452, 215–219 (2008).
Lister, R. et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133, 523–536 (2008).
The American Association for Cancer Research Human Epigenome Task Force European Union, Network of Excellence, Scientific Advisory Board Moving AHEAD with an international human epigenome project. Nature 454, 711–715 (2008).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Xi, Y. & Li, W. BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics 10, 232 (2009).
Coarfa, C. & Milosavljevic, A. Pash 2.0: scaleable sequence anchoring for next-generation sequencing technologies. Pac. Symp. Biocomput. 2008, 102–113 (2008).
Smith, A.D. et al. Updates to the RMAP short-read mapping software. Bioinformatics 25, 2841–2842 (2009).
Lin, H., Zhang, Z., Zhang, M.Q., Ma, B. & Li, M. ZOOM! Zillions of oligos mapped. Bioinformatics 24, 2431–2437 (2008).
Wang, T. et al. Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53. Proc. Natl. Acad. Sci. USA 104, 18613–18618 (2007).
Kunarso, G. et al. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat. Genet. 42, 631–634 (2010).
Pant, P.V.K. et al. Analysis of allelic differential expression in human white blood cells. Genome Res. 16, 331–339 (2006).
Pollard, K.S. et al. A genome-wide approach to identifying novel-imprinted genes. Hum. Genet. 122, 625–634 (2008).
Schalkwyk, L.C. et al. Allelic skewing of DNA methylation is widespread across the genome. Am. J. Hum. Genet. 86, 196–212 (2010).
Pick, M. et al. Clone- and gene-specific aberrations of parental imprinting in human induced pluripotent stem cells. Stem Cells 27, 2686–2690 (2009).
Arnaud, P. et al. Conserved methylation imprints in the human and mouse GRB10 genes with divergent allelic expression suggests differential reading of the same mark. Hum. Mol. Genet. 12, 1005–1019 (2003).
Li, N. et al. Whole genome DNA methylation analysis based on high throughput sequencing technology. Methods published online, doi: 10.1016/j.ymeth.2010.04.009 (27 April 2010).
Deng, J. et al. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat. Biotechnol. 27, 353–360 (2009).
Bourque, G. Transposable elements in gene regulation and in the evolution of vertebrate genomes. Curr. Opin. Genet. Dev. 19, 607–612 (2009).
Duhl, D.M., Vrieling, H., Miller, K.A., Wolff, G.L. & Barsh, G.S. Neomorphic agouti mutations in obese yellow mice. Nat. Genet. 8, 59–65 (1994).
Waterland, R.A. & Jirtle, R.L. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol. Cell. Biol. 23, 5293–5300 (2003).
Hellman, A. & Chess, A. Gene body-specific methylation on the active X chromosome. Science 315, 1141–1143 (2007).
Ludwig, T.E. et al. Feeder-independent culture of human embryonic stem cells. Nat. Methods 3, 637–646 (2006).
Gu, H. et al. Genome-scale DNA methylation mapping of clinical samples at single-nucleotide resolution. Nat. Methods 7, 133–136 (2010).
Smith, Z.D., Gu, H., Bock, C., Gnirke, A. & Meissner, A. High-throughput bisulfite sequencing in mammalian genomes. Methods 48, 226–232 (2009).
O'Geen, H., Frietze, S. & Farnham, P.J. Using ChIP-seq technology to identify targets of zinc finger transcription factors. Methods Mol. Biol. 649, 437–455 (2010).
Robertson, G. et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Methods 4, 651–657 (2007).
Blahnik, K.R. et al. Sole-Search: an integrated analysis program for peak detection and functional annotation using ChIP-seq data. Nucleic Acids Res. 38, e13 (2010).
Waterland, R.A., Lin, J., Smith, C.A. & Jirtle, R.L. Post-weaning diet affects genomic imprinting at the insulin-like growth factor 2 (Igf2) locus. Hum. Mol. Genet. 15, 705–716 (2006).
Shen, L., Guo, Y., Chen, X., Ahmed, S. & Issa, J.J. Optimizing annealing temperature overcomes bias in bisulfite PCR methylation analysis. Biotechniques 42, 48, 50, 52 passim (2007).
Grunau, C., Clark, S.J. & Rosenthal, A. Bisulfite genomic sequencing: systematic investigation of critical experimental parameters. Nucleic Acids Res 29, E65 (2001).
Acknowledgements
We would like to thank the US National Institutes of Health (NIH) Roadmap Epigenomics Program; sponsored by the National Institute on Drug Abuse (NIDA) and the National Institute of Environmental Health Sciences (NIEHS). J.F.C. and M.H. are supported by NIH grant 5U01ES017154-02. A. Milosavljevic is supported by NIH grant 5U01DA025956-02. A. Meissner and B.E.B. are supported by NIH grant 6U01ES017155-02. J.R.E. and B.R. are supported by NIH grant 5U01ES017166-02. R.P.N. was supported by NIH T32 CA108462-04 and F32CA141799. S.L.D. was supported by CIRM TB1-01190. S.D.F. was supported by NIH T32 CA108462-06. B.E.J. was supported by NIH T32 GM008568. M.A.M. is a Terry Fox Young Investigator and a Michael Smith Senior Research Scholar. We thank Z. Zhang and H. Li for modifying the ZOOM algorithm for bisulfite alignments.
Author information
Authors and Affiliations
Contributions
J.F.C., R.A.H., T.W., M.H., M.A.M. and A. Milosavljevic conceived and designed the experiments. R.P.N., C.H., S.L.D., B.E.J., S.D.F., Y.Z. and M.H. performed the MeDIP, MRE and bisulfite sequencing experiments. R.A.W. and X.Z. designed and performed pyrosequencing and data analyses. H.G., C.B., A.G. and A. Meissner9 performed and analyzed RRBS. L.E., H.O., P.J.F., B.E.B., C.B.E., R.D.H. and B.R. performed and analyzed Chip-seq experiments. R.L., M.P. and J.R.E. analyzed MethylC-seq data and performed Bowtie aligner testing. R.A.H., T.W., K.J.F., J.G., C.C., M.H., X.Z., A.D. and A.O. performed data analysis. T.W., T.B. and D.H. developed MeDIP and methyl-sensitive restriction enzyme scoring algorithms and performed coverage analyses including repetitive sequence analyses. Y.X., W.-Y.C., R.L., M.Q.Z. and W.L. compared bisulfite sequence aligners. J.F.C., R.A.H., M.H., T.W., R.P.N. and R.A.W. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 2, 4, 5 and 8 and Supplementary Figs. 1–18 (PDF 3818 kb)
Supplementary Table 1
Primer designs for bisulfite pyrosequencing. See Excel spreadsheet Supplementary_Table_1.xls. (XLS 33 kb)
Supplementary Table 3
Bisulfite data for Supplementary Figure 12. (XLS 118 kb)
Supplementary Table 6
Genome-wide catalogue of CpG island regions exhibiting overlapping MeDIP-seq (methylated) signals and MRE-seq (unmethylated) signals. (XLS 223 kb)
Supplementary Table 7
Validation of known and putative DMRs by bisulfite, PCR, cloning and sequencing. (XLS 250 kb)
Supplementary Table 9
Details of the comparison of genomic variation between pairs of assays to determine allele-specific epigenetic states. (XLS 409 kb)
Rights and permissions
About this article
Cite this article
Harris, R., Wang, T., Coarfa, C. et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat Biotechnol 28, 1097–1105 (2010). https://doi.org/10.1038/nbt.1682
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.1682
This article is cited by
-
Induction of double-strand breaks with the non-steroidal androgen receptor ligand flutamide in patients on androgen suppression: a study protocol for a randomized, double-blind prospective trial
Trials (2023)
-
BiocMAP: a Bioconductor-friendly, GPU-accelerated pipeline for bisulfite-sequencing data
BMC Bioinformatics (2023)
-
Broodstock nutritional programming differentially affects the hepatic transcriptome and genome-wide DNA methylome of farmed gilthead sea bream (Sparus aurata) depending on genetic background
BMC Genomics (2023)
-
Bisulfite-free mapping of DNA cytosine modifications: challenges and perspectives
Science China Chemistry (2023)
-
Metals Exposures and DNA Methylation: Current Evidence and Future Directions
Current Environmental Health Reports (2022)