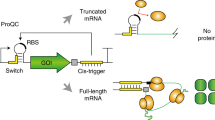
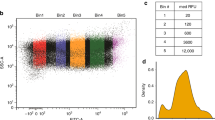
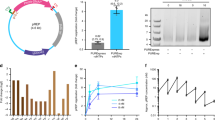
Abstract
Cell-free protein synthesis enables the rapid production and engineering of recombinant proteins. Existing cell-free systems differ substantially from each other with respect to efficiency, scalability and the ability to produce functional eukaryotic proteins. Here we describe species-independent translational sequences (SITS) that mediate efficient cell-free protein synthesis in multiple prokaryotic and eukaryotic systems, presumably through bypassing the early translation initiation factors. We use these leaders in combination with targeted suppression of the endogenous Leishmania tarentolae mRNAs to create a cell-free system based on this protozoan. The system can be directly programmed with unpurified PCR products, enabling rapid generation of large protein libraries and protein variants. L. tarentolae extract can produce up to 300 μg/ml of recombinant protein in 2 h. We further demonstrate that protein-protein and protein–small molecule interactions can be quantitatively analyzed directly in the translation mixtures using fluorescent (cross-) correlation spectroscopy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Nirenberg, M.W. & Matthaei, J.H. The dependence of cell-free protein synthesis in E. coli upon naturally occurring or synthetic polyribonucleotides. Proc. Natl. Acad. Sci. USA 47, 1588–1602 (1961).
Katzen, F., Chang, G. & Kudlicki, W. The past, present and future of cell-free protein synthesis. Trends Biotechnol. 23, 150–156 (2005).
Shimizu, Y. et al. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 19, 751–755 (2001).
Sawasaki, T., Morishita, R., Gouda, M.D. & Endo, Y. Methods for high-throughput materialization of genetic information based on wheat germ cell-free expression system. Methods Mol. Biol. 375, 95–106 (2007).
McCallum, C.D., Do, H., Johnson, A.E. & Frydman, J. The interaction of the chaperonin tailless complex polypeptide 1 (TCP1) ring complex (TRiC) with ribosome-bound nascent chains examined using photo-cross-linking. J. Cell Biol. 149, 591–602 (2000).
Ezure, T. et al. Cell-free protein synthesis system prepared from insect cells by freeze-thawing. Biotechnol. Prog. 22, 1570–1577 (2006).
He, M. & Wang, M.W. Arraying proteins by cell-free synthesis. Biomol. Eng. 24, 375–380 (2007).
Pestova, T.V. et al. Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl. Acad. Sci. USA 98, 7029–7036 (2001).
Merrick, W.C. Cap-dependent and cap-independent translation in eukaryotic systems. Gene 332, 1–11 (2004).
Lomakin, I.B., Hellen, C.U. & Pestova, T.V. Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol. Cell. Biol. 20, 6019–6029 (2000).
Shaloiko, L.A. et al. Effective non-viral leader for cap-independent translation in a eukaryotic cell-free system. Biotechnol. Bioeng. 88, 730–739 (2004).
Pelham, H.R. & Jackson, R.J. An efficient mRNA-dependent translation system from reticulocyte lysates. Eur. J. Biochem. 67, 247–256 (1976).
Korneeva, N.L., First, E.A., Benoit, C.A. & Rhoads, R.E. Interaction between the NH2-terminal domain of eIF4A and the central domain of eIF4G modulates RNA-stimulated ATPase activity. J. Biol. Chem. 280, 1872–1881 (2005).
Kozak, M. The scanning model for translation: an update. J. Cell Biol. 108, 229–241 (1989).
Kozak, M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J. Biol. Chem. 266, 19867–19870 (1991).
Pestova, T.V. & Kolupaeva, V.G. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 16, 2906–2922 (2002).
Ahn, B.Y. & Moss, B. Capped poly(A) leaders of variable lengths at the 5′ ends of vaccinia virus late mRNAs. J. Virol. 63, 226–232 (1989).
Bablanian, R., Goswami, S.K., Esteban, M., Banerjee, A.K. & Merrick, W.C. Mechanism of selective translation of vaccinia virus mRNAs: differential role of poly(A) and initiation factors in the translation of viral and cellular mRNAs. J. Virol. 65, 4449–4460 (1991).
Kamura, N., Sawasaki, T., Kasahara, Y., Takai, K. & Endo, Y. Selection of 5′-untranslated sequences that enhance initiation of translation in a cell-free protein synthesis system from wheat embryos. Bioorg. Med. Chem. Lett. 15, 5402–5406 (2005).
Berestovskaya, N.G. et al. Cotranslational formation of active photoprotein obelin in a cell-free translation system: direct ultrahigh sensitive measure of the translation course. Anal. Biochem. 268, 72–78 (1999).
Clayton, C. & Shapira, M. Post-transcriptional regulation of gene expression in trypanosomes and leishmanias. Mol. Biochem. Parasitol. 156, 93–101 (2007).
Breitling, R. et al. Non-pathogenic trypanosomatid protozoa as a platform for protein research and production. Protein Expr. Purif. 25, 209–218 (2002).
Niculae, A. et al. Isotopic labeling of recombinant proteins expressed in the protozoan host Leishmania tarentolae. Protein Expr. Purif. 48, 167–172 (2006).
Fritsche, C., Sitz, M., Wolf, M. & Pohl, H.D. Development of a defined medium for heterologous expression in Leishmania tarentolae. J. Basic Microbiol. 48, 488–495 (2008).
Leung, K.F., Baron, R. & Seabra, M.C. Thematic review series: lipid posttranslational modifications. geranylgeranylation of Rab GTPases. J. Lipid Res. 47, 467–475 (2006).
Rak, A. et al. Structure of the Rab7:REP-1 complex: insights into the mechanism of Rab prenylation and choroideremia disease. Cell 117, 749–760 (2004).
Nguyen, U.T. et al. Exploiting the substrate tolerance of farnesyltransferase for site-selective protein derivatization. ChemBioChem 8, 408–423 (2007).
Bacia, K. & Schwille, P. Practical guidelines for dual-color fluorescence cross-correlation spectroscopy. Nat. Protoc. 2, 2842–2856 (2007).
Oyama, R. et al. Protein-protein interaction analysis by C-terminally specific fluorescence labeling and fluorescence cross-correlation spectroscopy. Nucleic Acids Res. 34, e102 (2006).
Banaszynski, L.A., Liu, C.W. & Wandless, T.J. Characterization of the FKBP.rapamycin.FRB ternary complex. J. Am. Chem. Soc. 127, 4715–4721 (2005).
Warrens, A.N., Jones, M.D. & Lechler, R.I. Splicing by overlap extension by PCR using asymmetric amplification: an improved technique for the generation of hybrid proteins of immunological interest. Gene 186, 29–35 (1997).
Yakhnin, A.V., Vinokurov, L.M., Surin, A.K. & Alakhov, Y.B. Green fluorescent protein purification by organic extraction. Protein Expr. Purif. 14, 382–386 (1998).
Berrow, N.S. et al. A versatile ligation-independent cloning method suitable for high-throughput expression screening applications. Nucleic Acids Res. 35, e45 (2007).
Keese, M. et al. Quantitative imaging of apoptosis commitment in colorectal tumor cells. Differentiation 75, 809–818 (2007).
Rudolf, R. & Ulo, M. Diffusion of single molecules through a Gaussian laser beam in Proc. SPIE, 1921 (ed. Jouko, E.K.-T.) 239–248 (1993).
Acknowledgements
This work was supported in part by a Deutsche Forschungsgemeinschaft grant AL 484/5-4 and Heisenberg fellowship to K.A. U.T.T.N was supported by the predoctoral fellowship of Fonds der chemischen Industrie. We thank P. Lommerse for help with FCCS measurements and to V. Rajagopalan, M. Terbeck and A. Sander for excellent technical assistance. We are grateful to M. Gruen, R. Breitling, F. Seebeck and A. Goodall for critical reading of the manuscript and stimulating discussions and K. Browning (University of Texas at Austin) for a gift of reagents. We are very grateful to C. Clayton, S. Beverley, L. Shaloiko, I. Granowsky and P. Dittrich for support in the early stages of the project. We are indebted to R.S. Goody for long-term support.
Author information
Authors and Affiliations
Contributions
S.M. designed and performed research, O.K. designed and performed research, U.T.T.N designed and performed research and K.A. designed research and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
Kirill Alexandrov is a stakeholder in Jena Bioscience, which markets Leishmania tarentolae-based in vivo protein expression systems.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–16 and Supplementary Methods (PDF 1655 kb)
Rights and permissions
About this article
Cite this article
Mureev, S., Kovtun, O., Nguyen, U. et al. Species-independent translational leaders facilitate cell-free expression. Nat Biotechnol 27, 747–752 (2009). https://doi.org/10.1038/nbt.1556
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.1556
This article is cited by
-
Towards a generic prototyping approach for therapeutically-relevant peptides and proteins in a cell-free translation system
Nature Communications (2022)
-
Cell-Free Protein Synthesis: A Promising Option for Future Drug Development
BioDrugs (2020)
-
Identification of intracellular cavin target proteins reveals cavin-PP1alpha interactions regulate apoptosis
Nature Communications (2019)
-
Pathological mutations differentially affect the self-assembly and polymerisation of the innate immune system signalling adaptor molecule MyD88
BMC Biology (2018)
-
Protein structural biology using cell-free platform from wheat germ
Advanced Structural and Chemical Imaging (2018)