Abstract
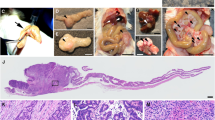
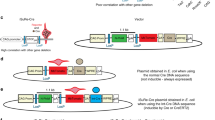
We describe a system that permits conditional mobilization of a Sleeping Beauty (SB) transposase allele by Cre recombinase to induce cancer specifically in a tissue of interest. To demonstrate its potential for developing tissue-specific models of cancer in mice, we limit SB transposition to the liver by placing Cre expression under the control of an albumin enhancer/promoter sequence and screen for hepatocellular carcinoma (HCC)–associated genes. From 8,060 nonredundant insertions cloned from 68 tumor nodules and comparative analysis with data from human HCC samples, we identify 19 loci strongly implicated in causing HCC. These encode genes, such as EGFR and MET, previously associated with HCC and others, such as UBE2H, that are potential new targets for treating this neoplasm. Our system, which could be modified to drive transposon-based insertional mutagenesis wherever tissue-specific Cre expression is possible, promises to enhance understanding of cancer genomes and identify new targets for therapeutic development.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Spradling, A.C. et al. Gene disruptions using P transposable elements: an integral component of the Drosophila genome project. Proc. Natl. Acad. Sci. USA 92, 10824–10830 (1995).
Bellen, H.J. et al. P-element-mediated enhancer detection: a versatile method to study development in Drosophila. Genes Dev. 3, 1288–1300 (1989).
Ivics, Z., Hackett, P.B., Plasterk, R.H. & Izsvak, Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell 91, 501–510 (1997).
Dupuy, A.J., Fritz, S. & Largaespada, D.A. Transposition and gene disruption in the male germline of the mouse. Genesis 30, 82–88 (2001).
Horie, K. et al. Efficient chromosomal transposition of a Tc1/mariner-like transposon Sleeping Beauty in mice. Proc. Natl. Acad. Sci. USA 98, 9191–9196 (2001).
Keng, V.W. et al. Region-specific saturation germline mutagenesis in mice using the Sleeping Beauty transposon system. Nat. Methods 2, 763–769 (2005).
Dupuy, A.J., Akagi, K., Largaespada, D.A., Copeland, N.G. & Jenkins, N.A. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature 436, 221–226 (2005).
Collier, L.S., Carlson, C.M., Ravimohan, S., Dupuy, A.J. & Largaespada, D.A. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature 436, 272–276 (2005).
Zambrowicz, B.P. et al. Disruption of overlapping transcripts in the ROSA beta geo 26 gene trap strain leads to widespread expression of beta-galactosidase in mouse embryos and hematopoietic cells. Proc. Natl. Acad. Sci. USA 94, 3789–3794 (1997).
Parkin, D.M., Bray, F., Ferlay, J. & Pisani, P. Global cancer statistics, 2002. CA Cancer J. Clin. 55, 74–108 (2005).
Llovet, J.M., Burroughs, A. & Bruix, J. Hepatocellular carcinoma. Lancet 362, 1907–1917 (2003).
Naugler, W.E. et al. Gender disparity in liver cancer due to sex differences in MyD88-dependent IL-6 production. Science 317, 121–124 (2007).
Bressac, B., Kew, M., Wands, J. & Ozturk, M. Selective G to T mutations of p53 gene in hepatocellular carcinoma from southern Africa. Nature 350, 429–431 (1991).
Buendia, M.A. Genetics of hepatocellular carcinoma. Semin. Cancer Biol. 10, 185–200 (2000).
Llovet, J.M. & Bruix, J. Molecular targeted therapies in hepatocellular carcinoma. Hepatology 48, 1312–1327 (2008).
Postic, C. et al. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J. Biol. Chem. 274, 305–315 (1999).
de Vries, A. et al. Targeted point mutations of p53 lead to dominant-negative inhibition of wild-type p53 function. Proc. Natl. Acad. Sci. USA 99, 2948–2953 (2002).
Rohde, F. et al. Expression of osteopontin, a target gene of de-regulated Wnt signaling, predicts survival in colon cancer. Int. J. Cancer 121, 1717–1723 (2007).
Calvisi, D.F., Factor, V.M., Loi, R. & Thorgeirsson, S.S. Activation of beta-catenin during hepatocarcinogenesis in transgenic mouse models: relationship to phenotype and tumor grade. Cancer Res. 61, 2085–2091 (2001).
Margulies, M. et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437, 376–380 (2005).
van Luenen, H.G., Colloms, S.D. & Plasterk, R.H. The mechanism of transposition of Tc3 in C. elegans. Cell 79, 293–301 (1994).
Vos, J.C., De Baere, I. & Plasterk, R.H. Transposase is the only nematode protein required for in vitro transposition of Tc1. Genes Dev. 10, 755–761 (1996).
Lampe, D.J., Churchill, M.E. & Robertson, H.M. A purified mariner transposase is sufficient to mediate transposition in vitro. EMBO J. 15, 5470–5479 (1996).
Chiang, D.Y. et al. Focal gains of VEGFA and molecular classification of hepatocellular carcinoma. Cancer Res. 68, 6779–6788 (2008).
Wangensteen, K.J. et al. A facile method for somatic, lifelong manipulation of multiple genes in the mouse liver. Hepatology 47, 1714–1724 (2008).
Bell, J.B. et al. Preferential delivery of the Sleeping Beauty transposon system to livers of mice by hydrodynamic injection. Nat. Protocols 2, 3153–3165 (2007).
Lee, T.K. et al. Regulation of angiogenesis by Id-1 through hypoxia-inducible factor-1alpha-mediated vascular endothelial growth factor up-regulation in hepatocellular carcinoma. Clin. Cancer Res. 12, 6910–6919 (2006).
Hirota, K., Murata, M., Itoh, T., Yodoi, J. & Fukuda, K. Redox-sensitive transactivation of epidermal growth factor receptor by tumor necrosis factor confers the NF-kappa B activation. J. Biol. Chem. 276, 25953–25958 (2001).
Arsura, M. et al. Transient activation of NF-kappaB through a TAK1/IKK kinase pathway by TGF-beta1 inhibits AP-1/SMAD signaling and apoptosis: implications in liver tumor formation. Oncogene 22, 412–425 (2003).
Chang, C.M. et al. A minor tyrosine phosphorylation site located within the CAIN domain plays a critical role in regulating tissue-specific transformation by erbB kinase. J. Virol. 69, 1172–1180 (1995).
Boerner, J.L., Danielsen, A. & Maihle, N.J. Ligand-independent oncogenic signaling by the epidermal growth factor receptor: v-ErbB as a paradigm. Exp. Cell Res. 284, 111–121 (2003).
Frederick, L., Wang, X.Y., Eley, G. & James, C.D. Diversity and frequency of epidermal growth factor receptor mutations in human glioblastomas. Cancer Res. 60, 1383–1387 (2000).
Landau, M., Fleishman, S.J. & Ben-Tal, N. A putative mechanism for downregulation of the catalytic activity of the EGF receptor via direct contact between its kinase and C-terminal domains. Structure 12, 2265–2275 (2004).
Sasaoka, T. et al. Involvement of ErbB2 in the signaling pathway leading to cell cycle progression from a truncated epidermal growth factor receptor lacking the C-terminal autophosphorylation sites. J. Biol. Chem. 271, 8338–8344 (1996).
Bekaii-Saab, T., Williams, N., Plass, C., Calero, M.V. & Eng, C. A novel mutation in the tyrosine kinase domain of ERBB2 in hepatocellular carcinoma. BMC Cancer 6, 278 (2006).
Buckley, A.F., Burgart, L.J., Sahai, V. & Kakar, S. Epidermal growth factor receptor expression and gene copy number in conventional hepatocellular carcinoma. Am. J. Clin. Pathol. 129, 245–251 (2008).
Rajkumar, T. & Gullick, W.J. The type I growth factor receptors in human breast cancer. Breast Cancer Res. Treat. 29, 3–9 (1994).
Lopes, L.F. & Bacchi, C.E. EGFR and gastrointestinal stromal tumor: an immunohistochemical and FISH study of 82 cases. Mod. Pathol. 20, 990–994 (2007).
Villanueva, A. et al. Pivotal role of mTOR signaling in hepatocellular carcinoma. Gastroenterology (2008).
Villanueva, A., Newell, P., Chiang, D.Y., Friedman, S.L. & Llovet, J.M. Genomics and signaling pathways in hepatocellular carcinoma. Semin. Liver Dis. 27, 55–76 (2007).
Tanabe, K.K. et al. Epidermal growth factor gene functional polymorphism and the risk of hepatocellular carcinoma in patients with cirrhosis. J. Am. Med. Assoc. 299, 53–60 (2008).
Philip, P.A. et al. Phase II study of erlotinib (OSI-774) in patients with advanced hepatocellular cancer. J. Clin. Oncol. 23, 6657–6663 (2005).
Salvi, A. et al. In vitro c-met inhibition by antisense RNA and plasmid-based RNAi down-modulates migration and invasion of hepatocellular carcinoma cells. Int. J. Oncol. 31, 451–460 (2007).
Takami, T. et al. Loss of hepatocyte growth factor/c-Met signaling pathway accelerates early stages of N-nitrosodiethylamine induced hepatocarcinogenesis. Cancer Res. 67, 9844–9851 (2007).
Su, G.H., Song, J.J., Repasky, E.A., Schutte, M. & Kern, S.E. Mutation rate of MAP2K4/MKK4 in breast carcinoma. Hum. Mutat. 19, 81 (2002).
Xin, W. et al. MAP2K4/MKK4 expression in pancreatic cancer: genetic validation of immunohistochemistry and relationship to disease course. Clin. Cancer Res. 10, 8516–8520 (2004).
Nakayama, K. et al. Homozygous deletion of MKK4 in ovarian serous carcinoma. Cancer Biol. Ther. 5, 630–634 (2006).
Iizuka, N. et al. Differential gene expression in distinct virologic types of hepatocellular carcinoma: association with liver cirrhosis. Oncogene 22, 3007–3014 (2003).
Moinzadeh, P., Breuhahn, K., Stutzer, H. & Schirmacher, P. Chromosome alterations in human hepatocellular carcinomas correlate with aetiology and histological grade–results of an explorative CGH meta-analysis. Br. J. Cancer 92, 935–941 (2005).
Zimmermann, U. et al. Chromosomal aberrations in hepatocellular carcinomas: relationship with pathological features. Hepatology 26, 1492–1498 (1997).
Zender, L. et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell 125, 1253–1267 (2006).
Wilber, A. et al. Messenger RNA as a source of transposase for sleeping beauty transposon-mediated correction of hereditary tyrosinemia type I. Mol. Ther. 15, 1280–1287 (2007).
Acknowledgements
The authors wish to thank Christine E. Nelson, Stefanie S. Breitbarth, Michelle K. Gleason and Geoff Hart for their excellent technical support; Jason B. Bell for performing the hydrodynamic injections; and Heidi Gruelich, Dana Farber Cancer Institute, for her kind gift pBabe-Puro-LTR-EGFR. We are also grateful to the Minnesota Supercomputing Institute for providing extensive computational resources (hardware and systems administration support) used to carry out the sequence analysis. A.V. is supported by a Sheila Sherlock fellowship from the European Association for the Study of the Liver. L.S.C. is supported by a 1K01CA122183-01 grant from the National Cancer Institute. N.G.C., N.A.J. and L.T. are supported by the Department of Health and Human Services, National Institutes of Health and the National Cancer Institute. J.M.L. is supported by the US National Institute of Diabetes and Digestive and Kidney Diseases (1R01DK076986-01), Spanish National Institute of Health (SAF-2007-61898) and Samuel Waxman Cancer Research Foundation. D.A.L. is supported by U01 CA84221 and R01 CA113636 grants from the National Cancer Institute.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Sleeping Beauty (SB) technology was exclusively licensed to Discovery Genomics (DGI, Minneapolis), cofounded by D.A.L., who has an equity interest in DGI and is an unpaid scientific advisor to DGI. DGI, which is pursuing the use of SB for human gene therapy, contributed neither money nor personnel to the present work. The work reported in this paper is not related to DGI's future plans. The University of Minnesota has filed a patent application on the process of using transposons, such as SB, to find cancer genes in laboratory animals using a forward genetic approach like the one described here. D.A.L., L.S.C., A.J.D., N.A.J. and N.G.C. are named inventors on the patent.
Supplementary information
Supplementary Text and Figures
Figures 1–6, Tables 1–3, Methods (PDF 2700 kb)
Supplementary Data
8,060 non-redundant insertion sites (Excel file) (XLS 6327 kb)
Rights and permissions
About this article
Cite this article
Keng, V., Villanueva, A., Chiang, D. et al. A conditional transposon-based insertional mutagenesis screen for genes associated with mouse hepatocellular carcinoma. Nat Biotechnol 27, 264–274 (2009). https://doi.org/10.1038/nbt.1526
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.1526
This article is cited by
-
Mfap4: a promising target for enhanced liver regeneration and chronic liver disease treatment
npj Regenerative Medicine (2023)
-
Exploring liver cancer biology through functional genetic screens
Nature Reviews Gastroenterology & Hepatology (2021)
-
Know Your Model: A brief history of making mutant mouse genetic models
Lab Animal (2021)
-
EGFR activation limits the response of liver cancer to lenvatinib
Nature (2021)
-
The impact of endotrophin on the progression of chronic liver disease
Experimental & Molecular Medicine (2020)