Abstract
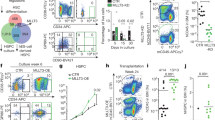
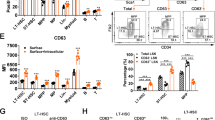
Hematopoietic stem cells (HSCs) are thought to divide infrequently based on their resistance to cytotoxic injury targeted at rapidly cycling cells1,2 and have been presumed to retain labels such as the thymidine analog 5-bromodeoxyuridine (BrdU). However, BrdU retention is neither a sensitive nor specific marker for HSCs3. Here we show that transient, transgenic expression of a histone 2B (H2B)–green fluorescent protein (GFP) fusion protein in mice has several advantages for label-retention studies over BrdU, including rapid induction of H2B-GFP in virtually all HSCs, higher labeling intensity and the ability to prospectively study label-retaining cells, which together permit a more precise analysis of division history. Mathematical modeling of H2B-GFP dilution in HSCs, identified with a stringent marker combination (L−K+S+CD48−CD150+)4, revealed unexpected heterogeneity in their proliferation rates and showed that ∼20% of HSCs divide at an extremely low rate (≤0.8–1.8% per day).
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Lerner, C. & Harrison, D.E. 5-Fluorouracil spares hemopoietic stem cells responsible for long-term repopulation. Exp. Hematol. 18, 114–118 (1990).
Van Zant, G. Studies of hematopoietic stem cells spared by 5-fluorouracil. J. Exp. Med. 159, 679–690 (1984).
Kiel, M.J. et al. Haematopoietic stem cells do not asymmetrically segregate chromosomes or retain BrdU. Nature 449, 238–242 (2007).
Kiel, M.J. et al. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell 121, 1109–1121 (2005).
Arai, F. et al. Tie2/angiopoietin-1 signaling regulates hematopoietic stem cell quiescence in the bone marrow niche. Cell 118, 149–161 (2004).
Zhang, J. et al. Identification of the haematopoietic stem cell niche and control of the niche size. Nature 425, 836–841 (2003).
Cotsarelis, G., Sun, T.T. & Lavker, R.M. Label-retaining cells reside in the bulge area of pilosebaceous unit: implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell 61, 1329–1337 (1990).
Kanda, T., Sullivan, K.F. & Wahl, G.M. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr. Biol. 8, 377–385 (1998).
Tumbar, T. et al. Defining the epithelial stem cell niche in skin. Science 303, 359–363 (2004).
Potten, C.S., Hume, W.J., Reid, P. & Cairns, J. The segregation of DNA in epithelial stem cells. Cell 15, 899–906 (1978).
Welm, B.E. et al. Sca-1(pos) cells in the mouse mammary gland represent an enriched progenitor cell population. Dev. Biol. 245, 42–56 (2002).
Urbanek, K. et al. Stem cell niches in the adult mouse heart. Proc. Natl. Acad. Sci. USA 103, 9226–9231 (2006).
Barker, N. et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 (2007).
Osawa, M., Hanada, K., Hamada, H. & Nakauchi, H. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science 273, 242–245 (1996).
Akala, O.O. et al. Long-term haematopoietic reconstitution by Trp53−/−p16Ink4a−/−p19Arf−/− multipotent progenitors. Nature 453, 228–232 (2008).
Kiel, M. J., Yilmaz, O. H. & Morrison, S. J. CD150- cells are transiently reconstituting multipotent progenitors with little or no stem cell activity. Blood 111, 4413–4414; author reply 4414–4415 (2008).
Balazs, A.B., Fabian, A.J., Esmon, C.T. & Mulligan, R.C. Endothelial protein C receptor (CD201) explicitly identifies hematopoietic stem cells in murine bone marrow. Blood 107, 2317–2321 (2006).
Chen, C.Z., Li, L., Li, M. & Lodish, H.F. The endoglin(positive) sca-1(positive) rhodamine(low) phenotype defines a near-homogeneous population of long-term repopulating hematopoietic stem cells. Immunity 19, 525–533 (2003).
Cheshier, S.H., Morrison, S.J., Liao, X. & Weissman, I.L. In vivo proliferation and cell cycle kinetics of long-term self-renewing hematopoietic stem cells. Proc. Natl. Acad. Sci. USA 96, 3120–3125 (1999).
Haug, J.S. et al. N-cadherin expression level distinguishes reserved versus primed states of hematopoietic stem cells. Cell Stem Cell 2, 367–379 (2008).
Jude, C.D., Gaudet, J.J., Speck, N.A. & Ernst, P. Leukemia and hematopoietic stem cells: balancing proliferation and quiescence. Cell Cycle 7, 586–591 (2008).
Cheng, T. et al. Hematopoietic stem cell quiescence maintained by p21cip1/waf1. Science 287, 1804–1808 (2000).
van Os, R. et al. A Limited role for p21Cip1/Waf1 in maintaining normal hematopoietic stem cell functioning. Stem Cells 25, 836–843 (2007).
Yamazaki, S. et al. Cytokine signals modulated via lipid rafts mimic niche signals and induce hibernation in hematopoietic stem cells. EMBO J. 25, 3515–3523 (2006).
Hock, H. et al. Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature 431, 1002–1007 (2004).
Zeng, H., Yucel, R., Kosan, C., Klein-Hitpass, L. & Moroy, T. Transcription factor Gfi1 regulates self-renewal and engraftment of hematopoietic stem cells. EMBO J. 23, 4116–4125 (2004).
Bradford, G.B., Williams, B., Rossi, R. & Bertoncello, I. Quiescence, cycling, and turnover in the primitive hematopoietic stem cell compartment. Exp. Hematol. 25, 445–453 (1997).
Sudo, K., Ema, H., Morita, Y. & Nakauchi, H. Age-associated characteristics of murine hematopoietic stem cells. J. Exp. Med. 192, 1273–1280 (2000).
Yilmaz, O.H., Kiel, M.J. & Morrison, S.J. SLAM family markers are conserved among hematopoietic stem cells from old and reconstituted mice and markedly increase their purity. Blood 107, 924–930 (2006).
Ema, H. et al. Quantification of self-renewal capacity in single hematopoietic stem cells from normal and Lnk-deficient mice. Dev. Cell 8, 907–914 (2005).
Beard, C., Hochedlinger, K., Plath, K., Wutz, A. & Jaenisch, R. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis 44, 23–28 (2006).
Hochedlinger, K., Yamada, Y., Beard, C. & Jaenisch, R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell 121, 465–477 (2005).
Hock, H. et al. Intrinsic requirement for zinc finger transcription factor Gfi-1 in neutrophil differentiation. Immunity 18, 109–120 (2003).
Hock, H. et al. Tel/Etv6 is an essential and selective regulator of adult hematopoietic stem cell survival. Genes Dev. 18, 2336–2341 (2004).
Miller, R. Survival Analysis (Wiley, NY; 1981).
Johnson, N.L., Kotz, S. & Balakrishnan, N. Continuous Uniform Distributions 2e (Wiley, NY; 1994).
Bates, D.M. & Watts, D.G. Nonlinear Regression Analysis and Its Application (Wiley, NY; 1988).
Acknowledgements
We thank Laura Prickett and Kat Folz-Donahue from the HSCI Flow-core at Massachusetts General Hospital (MGH) for their expert assistance, Geoffrey Wahl for providing the H2B-GFP cDNA, and Matthias Stadtfeld, Wenjun Guo and Ben Wittner for valuable input. We thank Linheng Li for the kind gift of biotinylated MNCD-2. We thank Mark Kiel and Sean Morrison for sharing unpublished results and for the gift of excised bone marrow of conditional N-cadherin mutant mice. We thank Glenn Radice for permission to use bone marrow from conditional N-cadherin mutant mice. This work was supported by a contribution from the Ellison Foundation to MGH startup funds for H.H., a seed grant by Harvard Stem Cell Institute to H.H. and by RO1CA122726. H.H. is the recipient of an ASH Scholar Award. K.H. was supported by the NIH Director's Innovator Award, the Harvard Stem Cell Institute, the Kimmel Foundation and the V Foundation. A.F. was the recipient of an MGH ECOR Fund for Medical Discovery Award.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Text and Figures
Figures 1–10 (PDF 1970 kb)
Rights and permissions
About this article
Cite this article
Foudi, A., Hochedlinger, K., Van Buren, D. et al. Analysis of histone 2B-GFP retention reveals slowly cycling hematopoietic stem cells. Nat Biotechnol 27, 84–90 (2009). https://doi.org/10.1038/nbt.1517
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.1517
This article is cited by
-
Elevated stress response marks deeply quiescent reserve cells of gastric chief cells
Communications Biology (2023)
-
A vertebral skeletal stem cell lineage driving metastasis
Nature (2023)
-
A multi-stem cell basis for craniosynostosis and calvarial mineralization
Nature (2023)
-
Mitochondrial metabolism as a potential therapeutic target in myeloid leukaemia
Leukemia (2022)
-
Fate mapping of hematopoietic stem cells reveals two pathways of native thrombopoiesis
Nature Communications (2022)