Abstract
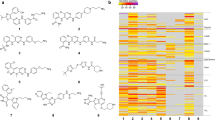
The aberrant activation of tyrosine kinases represents an important oncogenic mechanism, and yet the majority of such events remain undiscovered. Here we describe a bead-based method for detecting phosphorylation of both wild-type and mutant tyrosine kinases in a multiplexed, high-throughput and low-cost manner. With the aim of establishing a tyrosine kinase–activation catalog, we used this method to profile 130 human cancer lines. Follow-up experiments on the finding that SRC is frequently phosphorylated in glioblastoma cell lines showed that SRC is also activated in primary glioblastoma patient samples and that the SRC inhibitor dasatinib (Sprycel) inhibits viability and cell migration in vitro and tumor growth in vivo. Testing of dasatinib-resistant tyrosine kinase alleles confirmed that SRC is indeed the relevant target of dasatinib, which inhibits many tyrosine kinases. These studies establish the feasibility of tyrosine kinome–wide phosphorylation profiling and point to SRC as a possible therapeutic target in glioblastoma.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
References
Simon, M.P. et al. Deregulation of the platelet-derived growth factor B-chain gene via fusion with collagen gene COL1A1 in dermatofibrosarcoma protuberans and giant-cell fibroblastoma. Nat. Genet. 15, 95–98 (1997).
Beroukhim, R. et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc. Natl. Acad. Sci. USA 104, 20007–20012 (2007).
Heisterkamp, N., Stam, K., Groffen, J., de Klein, A. & Grosveld, G. Structural organization of the bcr gene and its role in the Ph' translocation. Nature 315, 758–761 (1985).
Tuveson, D.A. et al. STI571 inactivation of the gastrointestinal stromal tumor c-KIT oncoprotein: biological and clinical implications. Oncogene 20, 5054–5058 (2001).
Smolen, G.A. et al. Amplification of MET may identify a subset of cancers with extreme sensitivity to the selective tyrosine kinase inhibitor PHA-665752. Proc. Natl. Acad. Sci. USA 103, 2316–2321 (2006).
Nakamura, Y. et al. Constitutive activation of c-Met is correlated with c-Met overexpression and dependent on cell-matrix adhesion in lung adenocarcinoma cell lines. Cancer Sci. 99, 14–22 (2008).
Engelman, J.A. et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 316, 1039–1043 (2007).
Kita, Y.A. et al. NDF/heregulin stimulates the phosphorylation of Her3/erbB3. FEBS Lett. 349, 139–143 (1994).
Ueda, T. et al. Deletion of the carboxyl-terminal exons of K-sam/FGFR2 by short homology-mediated recombination, generating preferential expression of specific messenger RNAs. Cancer Res. 59, 6080–6086 (1999).
Desbois-Mouthon, C. et al. Dysregulation of glycogen synthase kinase-3beta signaling in hepatocellular carcinoma cells. Hepatology 36, 1528–1536 (2002).
Tibes, R. et al. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Mol. Cancer Ther. 5, 2512–2521 (2006).
Vescovi, A.L., Galli, R. & Reynolds, B.A. Brain tumour stem cells. Nat. Rev. Cancer 6, 425–436 (2006).
Libermann, T.A. et al. Amplification, enhanced expression and possible rearrangement of EGF receptor gene in primary human brain tumours of glial origin. Nature 313, 144–147 (1985).
Sugawa, N., Ekstrand, A.J., James, C.D. & Collins, V.P. Identical splicing of aberrant epidermal growth factor receptor transcripts from amplified rearranged genes in human glioblastomas. Proc. Natl. Acad. Sci. USA 87, 8602–8606 (1990).
Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068 (2008).
Lombardo, L.J. et al. Discovery of N-(2-chloro-6-methyl- phenyl)-2-(6-(4-(2-hydroxyethyl)-piperazin-1-yl)-2-methylpyrimidin-4-ylamino)thiazole-5-carboxamide (BMS-354825), a dual Src/Abl kinase inhibitor with potent antitumor activity in preclinical assays. J. Med. Chem. 47, 6658–6661 (2004).
Parsons, S.J. & Parsons, J.T. Src family kinases, key regulators of signal transduction. Oncogene 23, 7906–7909 (2004).
Angers-Loustau, A., Hering, R., Werbowetski, T.E., Kaplan, D.R. & Del Maestro, R.F. SRC regulates actin dynamics and invasion of malignant glial cells in three dimensions. Mol. Cancer Res. 2, 595–605 (2004).
Lund, C.V. et al. Reduced glioma infiltration in Src-deficient mice. J. Neurooncol. 78, 19–29 (2006).
Park, C.M. et al. Ionizing radiation enhances matrix metalloproteinase-2 secretion and invasion of glioma cells through Src/epidermal growth factor receptor-mediated p38/Akt and phosphatidylinositol 3-kinase/Akt signaling pathways. Cancer Res. 66, 8511–8519 (2006).
Phuong, L.K. et al. Use of a vaccine strain of measles virus genetically engineered to produce carcinoembryonic antigen as a novel therapeutic agent against glioblastoma multiforme. Cancer Res. 63, 2462–2469 (2003).
Carter, T.A. et al. Inhibition of drug-resistant mutants of ABL, KIT, and EGF receptor kinases. Proc. Natl. Acad. Sci. USA 102, 11011–11016 (2005).
Gorre, M.E. et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science 293, 876–880 (2001).
Shah, N.P. et al. Overriding imatinib resistance with a novel ABL kinase inhibitor. Science 305, 399–401 (2004).
Feinstein, A.R. Principles of Medical Statistics (Chapman & Hall / CRC Boca Raton, FL, 2002).
Efron, B. Nonparametric standard errors and confidence intervals. Can. J. Stat. 9, 139–158 (1981).
Rush, J. et al. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat. Biotechnol. 23, 94–101 (2005).
Rubin, J.B. et al. A small-molecule antagonist of CXCR4 inhibits intracranial growth of primary brain tumors. Proc. Natl. Acad. Sci. USA 100, 13513–13518 (2003).
Acknowledgements
We would like to thank Levi Garraway and Johnathan Fletcher for cell lines. We thank Julie Dang for her technical support on the pY419SRC IHC experiment. J.D. is supported by a fellowship from the Leukemia and Lymphoma Society and The Irving Family. I.K.M. is funded by the Brain Tumor Funders' Collaborative. The project has been funded in part with funds from the National Cancer Institute's (NCI) Initiative for Chemical Genetics, National Institutes of Health, under contract no. N01-CO-12400, and through the NCI's Integrative Cancer Biology Program.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3, Methods (PDF 288 kb)
Supplementary Table 1
Tyrosine phosphorylated proteins identified with IP-MS method in five cancer cell lines. (XLS 36 kb)
Supplementary Table 2
Capture antibodies and positive control samples used in Luminex immunosandwich assay. (XLS 35 kb)
Supplementary Table 3
Tyrosine phosphorylation levels on antibody validation samples. (XLS 75 kb)
Supplementary Table 4
Tyrosine phosphorylation levels of wild-type and dasatinib-resistant tyrosine kinase mutants measured by Luminex assay. (XLS 48 kb)
Supplementary Table 5
Tyrosine phosphorylation levels in six cell lines used for measuring technical and biological variations. (XLS 194 kb)
Supplementary Table 6
Tyrosine phosphorylation levels on cancer cell lines. (XLS 91 kb)
Supplementary Table 7
Tyrosine phosphorylation levels and genomic data on the primary glioma samples. (XLS 26 kb)
Supplementary Table 8
pY419SRC immunohistochemsitry on primary GBM tissue microarrays. (XLS 22 kb)
Supplementary Table 9
Dasatinib EC50 and SRC phosphorylation levels in cancer cell lines. (XLS 28 kb)
Supplementary Table 10
ORF amplification and mutagenesis primers for generating dasatinib-resistant mutants. (XLS 24 kb)
Rights and permissions
About this article
Cite this article
Du, J., Bernasconi, P., Clauser, K. et al. Bead-based profiling of tyrosine kinase phosphorylation identifies SRC as a potential target for glioblastoma therapy. Nat Biotechnol 27, 77–83 (2009). https://doi.org/10.1038/nbt.1513
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.1513
This article is cited by
-
Caspase-8 as a novel mediator linking Src kinase signaling to enhanced glioblastoma malignancy
Cell Death & Differentiation (2023)
-
Platelet-activating factor acetyl hydrolase IB2 dysregulated cell proliferation in ovarian cancer
Cancer Cell International (2021)
-
Multiplex glycan bead array for high throughput and high content analyses of glycan binding proteins
Nature Communications (2018)
-
Proinvasive extracellular matrix remodeling in tumor microenvironment in response to radiation
Oncogene (2018)
-
Upconversion Nanoparticles-Encoded Hydrogel Microbeads-Based Multiplexed Protein Detection
Nano-Micro Letters (2018)