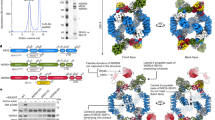
Abstract
Nutrients, such as amino acids and glucose, signal through the Rag GTPases to activate mTORC1. The GATOR1 protein complex—comprising DEPDC5, NPRL2 and NPRL3—regulates the Rag GTPases as a GTPase-activating protein (GAP) for RAGA; loss of GATOR1 desensitizes mTORC1 signalling to nutrient starvation. GATOR1 components have no sequence homology to other proteins, so the function of GATOR1 at the molecular level is currently unknown. Here we used cryo-electron microscopy to solve structures of GATOR1 and GATOR1–Rag GTPases complexes. GATOR1 adopts an extended architecture with a cavity in the middle; NPRL2 links DEPDC5 and NPRL3, and DEPDC5 contacts the Rag GTPase heterodimer. Biochemical analyses reveal that our GATOR1–Rag GTPases structure is inhibitory, and that at least two binding modes must exist between the Rag GTPases and GATOR1. Direct interaction of DEPDC5 with RAGA inhibits GATOR1-mediated stimulation of GTP hydrolysis by RAGA, whereas weaker interactions between the NPRL2–NPRL3 heterodimer and RAGA execute GAP activity. These data reveal the structure of a component of the nutrient-sensing mTORC1 pathway and a non-canonical interaction between a GAP and its substrate GTPase.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Efeyan, A., Comb, W. C. & Sabatini, D. M. Nutrient-sensing mechanisms and pathways. Nature 517, 302–310 (2015)
Shaw, R. J. & Cantley, L. C. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature 441, 424–430 (2006)
Jewell, J. L., Russell, R. C. & Guan, K.-L. Amino acid signalling upstream of mTOR. Nat. Rev. Mol. Cell Biol. 14, 133–139 (2013)
González, A. & Hall, M. N. Nutrient sensing and TOR signaling in yeast and mammals. EMBO J. 36, 397–408 (2017)
Saxton, R. A. & Sabatini, D. M. mTOR signaling in growth, metabolism, and disease. Cell 168, 960–976 (2017)
Schürmann, A., Brauers, A., Massmann, S., Becker, W. & Joost, H. G. Cloning of a novel family of mammalian GTP-binding proteins (RagA, RagBs, RagB1) with remote similarity to the Ras-related GTPases. J. Biol. Chem. 270, 28982–28988 (1995)
Hirose, E., Nakashima, N., Sekiguchi, T. & Nishimoto, T. RagA is a functional homologue of S. cerevisiae Gtr1p involved in the Ran/Gsp1-GTPase pathway. J. Cell Sci. 111, 11–21 (1998)
Sekiguchi, T., Hirose, E., Nakashima, N., Ii, M. & Nishimoto, T. Novel G proteins, Rag C and Rag D, interact with GTP-binding proteins, Rag A and Rag B. J. Biol. Chem. 276, 7246–7257 (2001)
Nakashima, N., Noguchi, E. & Nishimoto, T. Saccharomyces cerevisiae putative G protein, Gtr1p, which forms complexes with itself and a novel protein designated as Gtr2p, negatively regulates the Ran/Gsp1p G protein cycle through Gtr2p. Genetics 152, 853–867 (1999)
Sancak, Y. et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320, 1496–1501 (2008)
Inoki, K., Li, Y., Xu, T. & Guan, K.-L. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 17, 1829–1834 (2003)
Menon, S. et al. Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell 156, 771–785 (2014)
Li, Y., Inoki, K. & Guan, K. L. Biochemical and functional characterizations of small GTPase Rheb and TSC2 GAP activity. Mol. Cell. Biol. 24, 7965–7975 (2004)
Saito, K., Araki, Y., Kontani, K., Nishina, H. & Katada, T. Novel role of the small GTPase Rheb: its implication in endocytic pathway independent of the activation of mammalian target of rapamycin. J. Biochem. 137, 423–430 (2005)
Saucedo, L. J. et al. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat. Cell Biol. 5, 566–571 (2003)
Stocker, H. et al. Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat. Cell Biol. 5, 559–566 (2003)
Shen, K., Choe, A. & Sabatini, D. M. Intersubunit crosstalk in the Rag GTPase heterodimer enables mTORC1 to respond rapidly to amino acid availability. Mol. Cell 68, 552–565.e8 (2017)
Panchaud, N., Péli-Gulli, M.-P. & De Virgilio, C. Amino acid deprivation inhibits TORC1 through a GTPase-activating protein complex for the Rag family GTPase Gtr1. Sci. Signal. 6, ra42 (2013)
Bar-Peled, L. et al. A tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science 340, 1100–1106 (2013)
Petit, C. S., Roczniak-Ferguson, A. & Ferguson, S. M. Recruitment of folliculin to lysosomes supports the amino acid-dependent activation of Rag GTPases. J. Cell Biol. 202, 1107–1122 (2013)
Tsun, Z.-Y. et al. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol. Cell 52, 495–505 (2013)
Dibbens, L. M. et al. Mutations in DEPDC5 cause familial focal epilepsy with variable foci. Nat. Genet. 45, 546–551 (2013)
Ishida, S. et al. Mutations of DEPDC5 cause autosomal dominant focal epilepsies. Nat. Genet. 45, 552–555 (2013)
Wu, X. & Tu, B. P. Selective regulation of autophagy by the Iml1–Npr2–Npr3 complex in the absence of nitrogen starvation. Mol. Biol. Cell 22, 4124–4133 (2011)
Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9, 40 (2008)
Drozdetskiy, A., Cole, C., Procter, J. & Barton, G. J. JPred4: a protein secondary structure prediction server. Nucleic Acids Res. 43, W389–W394 (2015)
Levine, T. P. et al. Discovery of new Longin and Roadblock domains that form platforms for small GTPases in Ragulator and TRAPP-II. Small GTPases 4, 62–69 (2013)
Gong, R. et al. Crystal structure of the Gtr1p–Gtr2p complex reveals new insights into the amino acid-induced TORC1 activation. Genes Dev. 25, 1668–1673 (2011)
Jeong, J.-H. et al. Crystal structure of the Gtr1p(GTP)–Gtr2p(GDP) protein complex reveals large structural rearrangements triggered by GTP-to-GDP conversion. J. Biol. Chem. 287, 29648–29653 (2012)
Gibrat, J. F., Madej, T. & Bryant, S. H. Surprising similarities in structure comparison. Curr. Opin. Struct. Biol. 6, 377–385 (1996)
Shiozawa, K. et al. Structure of the N-terminal domain of PEX1 AAA-ATPase. Characterization of a putative adaptor-binding domain. J. Biol. Chem. 279, 50060–50068 (2004)
Ingelman, M., Bianchi, V. & Eklund, H. The three-dimensional structure of flavodoxin reductase from Escherichia coli at 1.7 Å resolution. J. Mol. Biol. 268, 147–157 (1997)
Qu, A. & Leahy, D. J. Crystal structure of the I-domain from the CD11a/CD18 (LFA-1, alpha L beta 2) integrin. Proc. Natl Acad. Sci. USA 92, 10277–10281 (1995)
Algret, R. et al. Molecular architecture and function of the SEA complex, a modulator of the TORC1 pathway. Mol. Cell. Proteomics 13, 2855–2870 (2014)
Kurzbauer, R. et al. Crystal structure of the p14/MP1 scaffolding complex: how a twin couple attaches mitogen-activated protein kinase signaling to late endosomes. Proc. Natl Acad. Sci. USA 101, 10984–10989 (2004)
Wolfson, R. L. et al. KICSTOR recruits GATOR1 to the lysosome and is necessary for nutrients to regulate mTORC1. Nature 543, 438–442 (2017)
Gu, X. et al. SAMTOR is an S-adenosylmethionine sensor for the mTORC1 pathway. Science 358, 813–818 (2017)
Scheffzek, K. et al. The Ras–RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science 277, 333–338 (1997)
Daumke, O., Weyand, M., Chakrabarti, P. P., Vetter, I. R. & Wittinghofer, A. The GTPase-activating protein Rap1GAP uses a catalytic asparagine. Nature 429, 197–201 (2004)
Acknowledgements
We thank all members of the Sabatini laboratory and T. Schwartz for insights; P. Abeyrathne, N. Grigorieff, R. Grant and C. Drennan for technical support; R. Saxton, M. Pacold and S. Shan for critical reading of the manuscript. This work was supported by grants from the NIH (R01 CA103866, R01 CA129105 and R37 AI047389) and Department of Defense (W81XWH-15-1-0230) to D.M.S., fellowship support from NSF (2016197106) to K.J.C. and from the Life Sciences Research Foundation to K.S., where he is a Pfizer Fellow. R.K.H., C.H. and Z.Y. were supported by the Howard Hughes Medical Institute. D.M.S. is an investigator of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
K.S. and D.M.S. initiated the project. K.S. purified the proteins and performed the biochemical characterization with input from K.J.C., M.L.V., L.C., A.B., and A.C. R.K.H., C.H. and Z.Y. determined the electron microscopy density maps for GATOR1 and GATOR1–Rag GTPases. K.S. and E.J.B. built the structural model. K.S., R.K.H., E.J.B., Z.Y. and D.M.S. wrote and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks D. Barford, K. Inoki and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 Structural determination and model building for the GATOR1 and GATOR1–Rag GTPases complex.
a, Gel filtration profiles for GATOR1 (red line) and GATOR1 + RAGA(T21N)–RAGC (orange line). The peak position for GATOR1 does not shift upon incubation with RAGA(T21N)–RAGC, which suggests that there is no direct binding between the two complexes. b, Coomassie blue stained SDS–PAGE analysis of the two peaks on the GATOR1 + RAGA(T21N)–RAGC elution profile. No co-elution is observed. Asterisk denotes a non-specific band that co-purifies with GATOR1. c, d, Raw cryo-EM images for GATOR1 (c) and the GATOR1–Rag GTPases complex (d). Discrete particles were clearly visualized under the microscope. Scale bars, 50 nm. e, f, 2D clustering of GATOR1 (e) and GATOR1–Rag GTPases (f). Yellow arrows in f point to the extra electron microscopy densities in comparison to e. * and § mark the particles shown in g. Scale bars, 5 nm. g, Direct comparison of particles from 2D clustering of GATOR1 and GATOR1–Rag GTPases. Extra electron microscopy densities for the Rag GTPases can be directly observed. h–j, Extracted regions from the electron microscopy density maps and the fitted structures for α-helical (h), β-strand (i) and loop (j) regions of DEPDC5. Secondary structures and bulky side chains can be unambiguously resolved at the current resolution. Data in a and b are representative of two independent experiments.
Extended Data Figure 2 Architecture of DEPDC5.
a, b, Two views of DEPDC5. The protein backbone is depicted in rainbow colours from the N terminus (blue) to the C terminus (red). Binding sites for NPRL2–NPRL3 and the Rag GTPases are marked. c, d, Structural model (c) and topological diagram (d) for DEPDC5 NTD. e, Lobe B of DEPDC5 NTD shares structural similarity to the NTD of the PEX1 AAA-ATPase. f–i, The SABA domain of DEPDC5 (f) shares topological similarity (g) to flavodoxin reductase (h) and CD11a I domain (i), which all contain ligand-binding sites (indicated by arrows). The SABA domain contains a β-sheet insertion formed by three strands. The three loops in the SABA domain of DEPDC5 that mediate the DEPDC5–NPRL2 interaction are coloured in red (loop A), orange (loop B), and blue (loop C), respectively, on the topological diagram. Nt, N terminus; Ct, C terminus.
Extended Data Figure 3 Architecture of the SHEN domain of DEPDC5.
a, Electron microscopy density map and structural model for the SHEN domain. b, Topological diagram for the SHEN domain. c, βH1 on linker S forms a continuous sheet with the β-strands on lobe B of the NTD, and positions itself between the NTD and the SABA domain. d, Electron microscopy density map and structural model for loop S. e, Loop S (purple) mediates interdomain contact with the SABA domain of DEPDC5, close to where the NPRL2–NPRL3 dimer binds to DEPDC5. f, βH2 (which we named the critical strip) of the SHEN domain directly contacts RAGA (pink). g, Electron microscopy density map and the atomic model for the critical strip.
Extended Data Figure 4 Architecture of the CTD of DEPDC5.
a, b, Structure (a) and topological diagram (b) for the CTD of DEPDC5. C, The CTD of DEPDC5 shows a pseudo two-fold rotational symmetry. Two lobes with similar folds can be seen.
Extended Data Figure 5 Architecture of NPRL2 and NPRL3.
a, Structural model of NPRL2. Contact surfaces with DEPDC5 and NPRL3 are indicated by arrows. A long linker connects the longin domain and the TINI domain, with electron microscopy density shown as mesh. The atomic model for this linker is shown in d. b, Longin domain of NPRL2. A standard longin domain from LST4 is shown for comparison. c, A strand-turn-strand motif (hairpin) is attached to the longin domain of NPRL2, which mediates partial interaction with DEPDC5. d, Electron microscopy density map and atomic model for the linker connecting the longin domain and the TINI domain (see the electron microscopy density in a). e, Structural model for the CTD of NPRL2. f, Structural model for NPRL3. Contact surfaces with NPRL2 are indicated by arrows. g, Longin domain of NPRL3 and its overlap with the longin domain of NPRL2. h, Structural model for the TINI domain of NPRL3 that connects its longin domain with the CTDs. i, Structural model for the intermediary (INT) domain of NPRL3. j, Structural model for the CTD of NPRL3. k–m, Interactions between NPRL2 and NPRL3. Three contact surfaces were identified that mediate the interactions between NPRL2 and NPRL3: the longin domains of NPRL2 and NPRL3 (k), the TINI domain of NPRL2 and CTD of NPRL3 (l), and the CTD of NPRL2 and the INT domain of NPRL3 (m).
Extended Data Figure 6 Architecture of the Rag GTPase heterodimer.
a, NBDs of RAGA (pink) and RAGC (cyan) overlap with those of Gtr1p and Gtr2p (grey). b, Extra electron microscopy density can be observed in the nucleotide-binding pocket of RAGA, into which GppNHp can be fitted. c, The CRD of RAGA and RAGC tightly dimerize with one another. The dimerized roadblock domains from Gtr1p–Gtr2p and p14–MP1 are shown for comparison. d, Global conformation of the Rag GTPase heterodimer in comparison to the two crystal structures of Gtr1p–Gtr2p. RAGA and Gtr1p are aligned. Rotational movement of the NBD of RAGC is illustrated, and compared with the NBD of Gtr2p in the direction of αN5. The NBDs of the Rag GTPases rotate further away from one another, even when compared with the open conformation of Gtr1p–Gtr2p (top).
Extended Data Figure 7 GATOR1 orchestrates amino acid signalling on the lysosomal surface.
a, Co-immunoprecipitation of GATOR2, KICSTOR and SAMTOR components by overexpressed GATOR1 in HEK293T cells. DEPDC5 by itself is sufficient to pull down endogenous KICSTOR components and SAMTOR. NPRL3 is necessary to pull down GATOR2 components. b, Co-immunoprecipitation of endogenous GATOR2 components by overexpressed NPRL2–NPRL3 in cells lacking DEPDC5. The NPRL2–NPRL3 dimer is sufficient to pull down GATOR2, and additional DEPDC5 causes no further effect. c, Signals from amino acids and metabolites are transmitted to GATOR1 through various routes. Data in a and b are representative of two independent experiments.
Extended Data Figure 8 Interactions between DEPDC5 and NPRL2.
a, b, Large contact surfaces between DEPDC5 (green) and NPRL2 (yellow) are observed from the electron microscopy density map and structural models. c, d, Surface residues on NPRL2 (c) and DEPDC5 (d) participate in mediating interactions between the two proteins, identified by ‘InterfaceResidue’ script in Pymol. e–g, Loops A (e), B (f) and C (g) on DEPDC5 directly contact NPRL2.
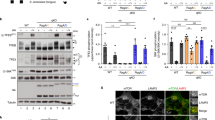
Extended Data Figure 9 In vitro characterization of the GAP mechanism of GATOR1.
a, Gel filtration profiles for DEPDC5(Y775A)–NPRL2–NPRL3 (blue line) and DEPDC5(Y775A)–NPRL2–NPRL3 + RAGA–RAGC(S75N) in the absence (orange line) or presence (red line) of the crosslinker glutaraldehyde. Peak A denotes the species eluted at the large molecular weight region. b, Coomassie blue stained SDS–PAGE analysis of peak A. Direct binding is only observed in the presence of glutaraldehyde. Asterisk denotes a non-specific band that co-purifies with GATOR1. c, No extra electron microscopy density can be observed near the NBD of RAGA. d, GATOR1 variants visualized by SDS–PAGE followed by Coomassie blue staining. Asterisk denotes a non-specific band that co-purifies with GATOR1. e, Scheme for measuring stimulated GTP hydrolysis by GATOR1 in a multiple turnover setup. An excess amount of Rag GTPases singly loaded with GTP was incubated with fixed amount of GATOR1. The hydrolysis reaction was traced and quantified. f, Stimulated GTP hydrolysis by wild-type GATOR1 shows a biphasic behaviour in reaction kinetics. As an increasing amount of the Rag GTPases was included in the reaction, a small plateau of observed rate constant (kobsd) was first observed at a lower concentration (inset). Such biphasic behaviour indicates that two binding modes exist in the wild-type GATOR1: one with higher affinity to the Rag GTPases but lower GAP activity, the other with lower affinity but higher GAP activity. A representative dataset is shown in this panel, and the statistics are summarized below. g, Stimulated GTP hydrolysis by a GATOR1 mutant that is defective in Rag GTPases binding eliminates the initial phase. DEPDC5(Y775A)–NPRL2–NPRL3 is defective in stable Rag GTPases binding because it lacks the docking site (an intact critical strip) for the Rag GTPases. Consequentially, the inhibitory mode diminishes (inset), leaving a single phase corresponding to the GAP mode in reaction kinetics. A representative dataset is shown, and the statistics are summarized below. h, Summary of kinetic parameters for the multiple turnover GAP activity shown in f and g. Mean ± s.d. of two to three independent experiments is reported. i, A stimulated-chase assay to characterize the inhibition mechanism of DEPDC5. Wild-type GATOR1 was first added to bind the Rag GTPases with its inhibitory mode. Extra NPRL2–NPRL3 was then included in the reaction as a chase. We reasoned that if DEPDC5 sequesters the NBD of RAGA, no further stimulation should be observed; if DEPDC5 simply prevents NPRL2–NPRL3 from accessing the NBD of RAGA, we should observe additional stimulation because there is no DEPDC5 to inhibit the extra NPRL2–NPRL3. j, Further stimulation is observed in the presence of additional NPRL2–NPRL3, as reflected by the faster hydrolysis rate (steeper slope), suggesting DEPDC5 inhibits NPRL2–NPRL3 in cis. Data in a, b, d and j are representative of two independent experiments.
Extended Data Figure 10 In vivo characterization of the GAP mechanism of GATOR1.
a, Interaction between NPRL2–NPRL3 and the Rag GTPases is enhanced by wild-type DEPDC5 but not mutant P, which is defective in binding to the Rag GTPases. W, wild-type DEPDC5; P, mutant P. Asterisk denotes a non-specific band. b, Amino acid availability regulates the interaction between NPRL2–NPRL3 and the Rag GTPases in cells lacking DEPDC5. Higher amount of Rag GTPases co-immunoprecipitates with NPRL2–NPRL3 in the absence of amino acids. c, Loss of regulated interaction between NPRL2–NPRL3 and the Rag GTPases in cells lacking DEPDC5 and MIOS. No difference is observed when GATOR2, the receptor for amino acid signals, is knocked out. d, Expression of a Dedpc5 mutant that is defective in Rag GTPases binding has no effect in NPRL2-null cells, in sharp contrast to the result in Fig. 6e. Data in a–d are representative of two independent experiments.
Supplementary information
Supplementary Figure
This file contains Supplementary Figure 1 which shows the uncropped gel pictures for all the western blot analyses in the article. (PDF 1254 kb)
Supplementary Table
This file contains Supplementary Table 1 which shows the EM data collection and model refinement. (PDF 99 kb)
Supplementary Information
This file contains Supplementary Material and Methods. (PDF 320 kb)
Rights and permissions
About this article
Cite this article
Shen, K., Huang, R., Brignole, E. et al. Architecture of the human GATOR1 and GATOR1–Rag GTPases complexes. Nature 556, 64–69 (2018). https://doi.org/10.1038/nature26158
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature26158
This article is cited by
-
Lysosomes as coordinators of cellular catabolism, metabolic signalling and organ physiology
Nature Reviews Molecular Cell Biology (2024)
-
Targeting the biology of aging with mTOR inhibitors
Nature Aging (2023)
-
The molecular basis of nutrient sensing and signalling by mTORC1 in metabolism regulation and disease
Nature Reviews Molecular Cell Biology (2023)
-
AMPK-dependent phosphorylation of the GATOR2 component WDR24 suppresses glucose-mediated mTORC1 activation
Nature Metabolism (2023)
-
Functional characterization of novel NPRL3 mutations identified in three families with focal epilepsy
Science China Life Sciences (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.