Abstract
The TGFβ pathway has essential roles in embryonic development, organ homeostasis, tissue repair and disease1,2. These diverse effects are mediated through the intracellular effectors SMAD2 and SMAD3 (hereafter SMAD2/3), whose canonical function is to control the activity of target genes by interacting with transcriptional regulators3. Therefore, a complete description of the factors that interact with SMAD2/3 in a given cell type would have broad implications for many areas of cell biology. Here we describe the interactome of SMAD2/3 in human pluripotent stem cells. This analysis reveals that SMAD2/3 is involved in multiple molecular processes in addition to its role in transcription. In particular, we identify a functional interaction with the METTL3–METTL14–WTAP complex, which mediates the conversion of adenosine to N6-methyladenosine (m6A) on RNA4. We show that SMAD2/3 promotes binding of the m6A methyltransferase complex to a subset of transcripts involved in early cell fate decisions. This mechanism destabilizes specific SMAD2/3 transcriptional targets, including the pluripotency factor gene NANOG, priming them for rapid downregulation upon differentiation to enable timely exit from pluripotency. Collectively, these findings reveal the mechanism by which extracellular signalling can induce rapid cellular responses through regulation of the epitranscriptome. These aspects of TGFβ signalling could have far-reaching implications in many other cell types and in diseases such as cancer5.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Wu, M. Y. & Hill, C. S. TGFβ superfamily signaling in embryonic development and homeostasis. Dev. Cell 16, 329–343 (2009)
Oshimori, N . & Fuchs, E. The harmonies played by TGFβ in stem cell biology. Cell Stem Cell 11, 751–764 (2012)
Gaarenstroom, T. & Hill, C. S. TGFβ signaling to chromatin: how SMADs regulate transcription during self-renewal and differentiation. Semin. Cell Dev. Biol. 32, 107–118 (2014)
Heyn, H. & Esteller, M. An adenine code for DNA: a second life for N6-methyladenine. Cell 161, 710–713 (2015)
Pickup, M., Novitskiy, S. & Moses, H. L. The roles of TGFβ in the tumour microenvironment. Nat. Rev. Cancer 13, 788–799 (2013)
Vallier, L., Reynolds, D. & Pedersen, R. A. NODAL inhibits differentiation of human embryonic stem cells along the neuroectodermal default pathway. Dev. Biol. 275, 403–421 (2004)
Vallier, L., Alexander, M. & Pedersen, R. A. Activin/NODAL and FGF pathways cooperate to maintain pluripotency of human embryonic stem cells. J. Cell Sci. 118, 4495–4509 (2005)
James, D., Levine, A. J., Besser, D. & Hemmati-Brivanlou, A. TGFβ/activin/NODAL signaling is necessary for the maintenance of pluripotency in human embryonic stem cells. Development 132, 1273–1282 (2005)
Smith, J. R. et al. Inhibition of activin/NODAL signaling promotes specification of human embryonic stem cells into neuroectoderm. Dev. Biol. 313, 107–117 (2008)
Bertero, A. et al. Activin/NODAL signaling and NANOG orchestrate human embryonic stem cell fate decisions by controlling the H3K4me3 chromatin mark. Genes Dev. 29, 702–717 (2015)
D’Amour, K. A. et al. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat. Biotechnol. 23, 1534–1541 (2005)
Vallier, L. et al. Signaling pathways controlling pluripotency and early cell fate decisions of human induced pluripotent stem cells. Stem Cells 27, 2655–2666 (2009)
Teo, A. K. et al. Pluripotency factors regulate definitive endoderm specification through eomesodermin. Genes Dev. 25, 238–250 (2011)
Kubo, A. et al. Development of definitive endoderm from embryonic stem cells in culture. Development 131, 1651–1662 (2004)
Ke, S. et al. A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation. Genes Dev. 29, 2037–2053 (2015)
Ke, S. et al. m6A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev. 31, 990–1006 (2017)
Dominissini, D. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 (2012)
Meyer, K. D. et al. 5′ UTR m6A promotes cap-independent translation. Cell 163, 999–1010 (2015)
Meyer, K. D. et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646 (2012)
Barbieri, I. et al. Promoter-bound METTL3 maintains myeloid leukaemia by m6A-dependent translation control. Nature 552, 126–131 (2017)
Vu, L. P. et al. The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med. 23, 1369–1376 (2017)
Geula, S. et al. Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 347, 1002–1006 (2015)
Batista, P. J. et al. m6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 15, 707–719 (2014)
Schwartz, S. et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Reports 8, 284–296 (2014)
Wang, X. et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120 (2014)
Bartosovic, M. et al. N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3′-end processing. Nucleic Acids Res. 45, 11356–11370 (2017)
Bertero, A. et al. Optimized inducible shRNA and CRISPR/Cas9 platforms for in vitro studies of human development using hPSCs. Development 143, 4405–4418 (2016)
Vallier, L. et al. Activin/NODAL signalling maintains pluripotency by controlling NANOG expression. Development 136, 1339–1349 (2009)
Mendjan, S. et al. NANOG and CDX2 pattern distinct subtypes of human mesoderm during exit from pluripotency. Cell Stem Cell 15, 310–325 (2014)
Brown, S. et al. Activin/NODAL signaling controls divergent transcriptional networks in human embryonic stem cells and in endoderm progenitors. Stem Cells 29, 1176–1185 (2011)
Yusa, K. et al. Targeted gene correction of α1-antitrypsin deficiency in induced pluripotent stem cells. Nature 478, 391–394 (2011)
Vallier, L. Serum-free and feeder-free culture conditions for human embryonic stem cells. Methods Mol. Biol. 690, 57–66 (2011)
Touboul, T. et al. Generation of functional hepatocytes from human embryonic stem cells under chemically defined conditions that recapitulate liver development. Hepatology 51, 1754–1765 (2010)
Vallier, L. et al. Early cell fate decisions of human embryonic stem cells and mouse epiblast stem cells are controlled by the same signalling pathways. PLoS One 4, e6082 (2009)
Moffat, J. et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124, 1283–1298 (2006)
Pawlowski, M. et al. Inducible and deterministic forward programming of human pluripotent stem cells into neurons, skeletal myocytes, and oligodendrocytes. Stem Cell Reports 8, 803–812 (2017)
Hubner, N. C. & Mann, M. Extracting gene function from protein-protein interactions using quantitative BAC interactomics (QUBIC). Methods 53, 453–459 (2011)
Rappsilber, J., Mann, M. & Ishihama, Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 (2007)
Boersema, P. J., Raijmakers, R., Lemeer, S., Mohammed, S. & Heck, A. J. R. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc. 4, 484–494 (2009)
Hubner, N. C., Nguyen, L. N., Hornig, N. C. & Stunnenberg, H. G. A quantitative proteomics tool to identify DNA–protein interactions in primary cells or blood. J. Proteome Res. 14, 1315–1329 (2015)
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008)
Hubner, N. C. et al. Quantitative proteomics combined with BAC transgeneomics reveals in vivo protein interactions. J. Cell Biol. 189, 739–754 (2010)
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003)
Chen, E. Y. et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 (2013)
Dominissini, D., Moshitch-Moshkovitz, S., Salmon-Divon, M., Amariglio, N. & Rechavi, G. Transcriptome-wide mapping of N6-methyladenosine by m6A-seq based on immunocapturing and massively parallel sequencing. Nat. Protoc. 8, 176–189 (2013)
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014)
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013)
Cui, X. et al. MeTDiff: a novel differential RNA methylation analysis for MeRIP–seq data. IEEE/ACM Trans. Comput. Biol. Bioinforma. PP, 1 (2015)
Saletore, Y. et al. The birth of the epitranscriptome: deciphering the function of RNA modifications. Genome Biol. 13, 175 (2012)
Li, X., Xiong, X. & Yi, C. Epitranscriptome sequencing technologies: decoding RNA modifications. Nat. Methods 14, 23–31 (2017)
Zhang, Y. et al. Model-based analysis of ChIP–seq (MACS). Genome Biol. 9, R137 (2008)
Madrigal, P. fCCAC: functional canonical correlation analysis to evaluate covariance between nucleic acid sequencing datasets. Bioinformatics 33, 746–748 (2017)
Wang, L., Wang, S. & Li, W. RSeQC: quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185 (2012)
Uyar, B. et al. RCAS: an RNA centric annotation system for transcriptome-wide regions of interest. Nucleic Acids Res. 45, e91 (2017)
Bailey, T. L. DREME: motif discovery in transcription factor ChIP–seq data. Bioinformatics 27, 1653–1659 (2011)
Down, T. A., Piipari, M. & Hubbard, T. J. P. Dalliance: interactive genome viewing on the web. Bioinformatics 27, 889–890 (2011)
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014)
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014)
Conesa, A. et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 17, 13 (2016)
Heger, A., Webber, C., Goodson, M., Ponting, C. P. & Lunter, G. GAT: a simulation framework for testing the association of genomic intervals. Bioinformatics 29, 2046–2048 (2013)
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010)
Meng, J., Cui, X., Rao, M. K., Chen, Y. & Huang, Y. Exome-based analysis for RNA epigenome sequencing data. Bioinformatics 29, 1565–1567 (2013)
Trapnell, C. et al. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010)
Joshi, N. & Fass, J. Sickle: a sliding-window, adaptive, quality-based trimming tool for FastQ files v.1.33 https://github.com/najoshi/sickle (2011)
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012)
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009)
Andrews, S. SeqMonk: A tool to visualise and analyse high throughput mapped sequence data https://www.bioinformatics.babraham.ac.uk/projects/seqmonk/ (2014)
Smyth, G. K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, e3 (2004)
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25, 402–408 (2001)
Harrold, S., Genovese, C., Kobrin, B., Morrison, S. L. & Milcarek, C. A comparison of apparent mRNA half-life using kinetic labeling techniques vs decay following administration of transcriptional inhibitors. Anal. Biochem. 198, 19–29 (1991)
Acknowledgements
We thank Cambridge Genomic Services for help with next-generation sequencing. This work was supported by the European Research Council starting grant ‘Relieve IMDs’ (L.V., S.B., A.B., P.M.); the Cambridge University Hospitals National Institute for Health Research Biomedical Research Center (L.V., J.K., A.-S.L.); the Wellcome Trust PhD program (A.O., L.Y.); a British Heart Foundation PhD studentship (FS/11/77/39327 to A.B.); a Grant-in-Aid for JSPS Fellows (16J08005 to S.N.); and a core support grant from the Wellcome Trust and Medical Research Council to the Wellcome Trust–Medical Research Council Cambridge Stem Cell Institute.
Author information
Authors and Affiliations
Contributions
A.B. conceived the study, performed or contributed to most of the experiments, analysed data and wrote the manuscript with input from the other authors. S.B. contributed to study conception, performed co-immunoprecipitation, NeMeRIP and RNA-IP experiments, and analysed data. P.M., I.R.d.l.M, and C.S. analysed NeMeRIP–seq. A.O. performed PLA and co-immunoprecipitations and analysed RNA-seq data. D.O., L.Y. and J.K. assisted with hPSC gene editing and differentiation; N.C.H. performed quantitative proteomics and data analysis. A.-S.L., S.N. and R.G. assisted with hPSC culture. E.F. optimized NeMeRIP–seq libraries. J.U. contributed to study conception and supervision. H.G.S. supervised quantitative proteomics. S.M. contributed to study conception and supervision, and assisted with SMAD2/3 co-immunoprecipitation. L.V. conceived, supervised and supported the study, and wrote and provided final approval of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks C. Mason and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 Optimized co-immunoprecipitation protocol to define the SMAD2/3 interactome in hPSCs and early endoderm cells.
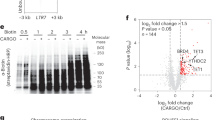
a, Western blots of SMAD2/3 or control (IgG) immunoprecipitations from nuclear extracts of hESCs following the co-IP1 or co-IP2 protocols. Input is 5% of the material used for immunoprecipitations. Results are representative of two independent experiments. For gel Source Data, see Supplementary Fig. 1. b, Scatter plots of the log2 ratios of label-free quantification (LFQ) intensities for proteins identified by quantitative mass spectrometry in SMAD2/3 co-immunoprecipitations compared with IgG negative control co-immunoprecipitations. The experiments were performed from nuclear extracts of hESCs. The SMAD2/3 and IgG negative control co-immunoprecipitations were differentially labelled after immunoprecipitation using the dimethyl method, followed by a combined run of the two samples in order to compare the abundance of specific peptides and identify enriched peptides. The values for technical dye-swap duplicates are plotted on different axes, and proteins whose enrichment was significant (significance B < 0.01) are shown in black and named. As a result of this comparison between the two co-immunoprecipitation protocols, co-IP2 was selected for further experiments (see Supplementary Discussion). c, Volcano plots of statistical significance against fold-change for proteins identified by label-free quantitative mass spectrometry in SMAD2/3 or IgG negative control immunoprecipitations in pluripotent hESCs or early endoderm (see Fig. 1a). The black lines indicate the threshold used to determine specific SMAD2/3 interactors, which are located to the right (n = 3 co-immunoprecipitations; one-tailed t-test: permutation-based FDR < 0.05). d, Selected results of the analysis described in c for SMAD2, SMAD3 and selected known bona fide SMAD2/3-binding partners (full results can be found in Supplementary Table 1). e, Mean label-free quantification (LFQ) intensity log2 ratios in endoderm (endo) and pluripotent cells (pluri) for all SMAD2/3 interactors. Differentially enriched proteins are shown as green and blue bars. f, Selected results from gene ontology (GO) enrichment analysis, and enrichment analysis for mouse phenotypes annotated in the MGI database. All putative SMAD2/3-interacting proteins were considered for this analysis (n = 89 proteins; Fisher’s exact test followed by Benjamini–Hochberg correction for multiple comparisons). For each term, its rank in the analysis, the adjusted P value, and the number of associated genes are reported.
Extended Data Figure 2 Functional characterization of transcriptional and epigenetic cofactors of SMAD2/3 in hPSCs.
a, Western blots of SMAD2/3 or control (IgG) immunoprecipitations from nuclear extracts of pluripotent hESCs (pluri) or hESCs differentiated into endoderm for 36 h (endo). Input is 5% of the material used for immunoprecipitations. Results are representative of two independent experiments. b, Schematic of the experimental approach for the generation of iKD hESC lines for SMAD2/3 cofactors. c, qPCR screening of iKD hESCs cultured in the absence (CTR) or presence (TET) of tetracycline for three days. Three distinct shRNAs were tested for each gene. Expression is normalized to the mean level in hESCs carrying negative control shRNAs (scrambled (SCR) or against B2M) and cultured in the absence of tetracycline. The mean is indicated, n = 2 independent clonal pools. Note than for the B2M shRNA only the scrambled shRNA was used as negative control. shRNAs selected for further experiments are circled. d, Phase-contrast images of iKD hESCs expressing the indicated shRNAs (sh) and cultured in the presence of tetracycline for six days to induce knockdown. Scale bars, 400 μm. Results are representative of two independent experiments. e, Immunofluorescence for the pluripotency factor NANOG in iKD hESCs for the indicated genes cultured in the absence (CTR) or presence of tetracycline (TET) for six days. Scale bars, 400 μm. Results are representative of two independent experiments. f, Heat map summarizing qPCR analyses of iKD hESCs cultured as in e. log2 fold-changes (FC) are compared to scrambled control (n = 2 clonal pools). Germ-layer markers are grouped in boxes: green, endoderm; red, mesoderm; blue, neuroectoderm.
Extended Data Figure 3 Functional characterization of transcriptional and epigenetic cofactors of SMAD2/3 during endoderm differentiation.
a, qPCR validation of iKD hESCs in pluripotent cells (PLURI) or following endoderm differentiation (ENDO). Pluripotent cells were cultured in the absence (CTR) or presence (TET) of tetracycline for six days. For endoderm differentiation, tetracycline treatment was initiated in undifferentiated hESCs for three days in order to ensure gene knockdown at the start of endoderm specification, and was then maintained during differentiation (three days). For each gene, the shRNA resulting in the strongest level of knockdown in hPSCs was selected (refer to Extended Data Fig. 2). Expression is normalized to the mean level in pluripotent hESCs carrying scrambled control shRNA and cultured in the absence of tetracycline. The mean is indicated, n = 2 independent clonal pools. b, Immunofluorescence of the endoderm marker SOX17 following endoderm differentiation of iKD hESCs expressing the indicated shRNAs and cultured as described in a. Scale bars, 400 μm. Results are representative of two independent experiments. c, qPCR following endoderm differentiation of iKD hESCs. The mean is indicated, n = 2 independent clonal pools. d, Table summarizing the phenotypic results presented in Extended Data Fig. 2 and in this figure. E, endoderm; N, neuroectoderm; M, mesoderm.
Extended Data Figure 4 Mechanistic insights into the functional interaction between SMAD2/3 and the m6A methyltransferase complex.
a–c, Western blots of SMAD2/3, METTL3, METTL14 or control (IgG) immunoprecipitations from nuclear extracts of hESCs (a, c) or hiPSCs (b). Input is 5% of the material used for immunoprecipitations. In c, immunoprecipitations were performed from hPSCs maintained in the presence of activin or treated for 1 h with the activin–NODAL signalling inhibitor SB431542. Results are representative of three (a) or two (b, c) independent experiments. d, qPCR validation of hESCs constitutively overexpressing NANOG (NANOG OE) following gene targeting of the AAVS1 locus with pAAV-Puro_CAG-NANOG. Parental wild-type H9 hESCs (H9) were analysed as negative controls. Cells were cultured in the presence of activin or treated with SB431542 for the indicated times. The mean is shown, n = 2 cultures. NANOG-overexpressing cells are resistant to downregulation of NANOG following inhibiton of activin–NODAL signalling. e, RNA immunoprecipitation experiments for WTAP, SMAD2/3 or IgG control in NANOG-overexpressing hESCs maintained in the presence of activin or treated for 2 h with SB431542. Enrichment of the indicated transcripts was measured by qPCR and expressed relative to background levels observed in control IgG RNA immunoprecipitations in the presence of activin. RPLP0 was tested as a negative control transcript. Mean ± s.e.m., n = 3 cultures. Significance of differences from activin (left) or IgG (right) RIP was tested by two-way ANOVA with post hoc Holm–Sidak comparisons. f, ChIP–qPCR in hESCs for ChIP against the indicated proteins or the negative control ChIP (IgG). qPCR was performed for validated genomic SMAD2/3-binding sites associated with the indicated genes10,30. hESCs were cultured in the presence of activin or treated for 2 h with SB431542. Enrichment is normalized against background binding observed with IgG ChIP. The mean is shown, n = 2 technical replicates. Results are representative of three independent experiments.
Extended Data Figure 5 Monitoring changes in m6A deposition that are rapidly induced by inhibition of activin–NODAL signalling.
a, b, MeRIP–qPCR results from purified mRNA, total cellular RNA or cellular RNA species separated by nuclear and cytoplasmic subcellular fractionation. hESCs were cultured in pluripotency-maintaining conditions containing activin or in conditions in which activin–NODAL signalling was inhibited for 2 h with SB431542. IgG MeRIP experiments were performed as negative controls. The mean is indicated, n = 2 technical replicates. Differences between activin- and SB431542-treated cells were observed only in the nuclear-enriched fraction. Therefore, the nuclear-enriched MeRIP protocol (NeMeRIP) was used for subsequent experiments (refer to the Supplementary Discussion). Results are representative of two independent experiments. c, Overlap with the indicated genomic features of m6A peaks identified by NeMeRIP–seq using two different bioinformatics pipelines in which peak calling was performed using MetDiff or MACS2. For each pipeline, the analyses were performed on the union of peaks identified from data obtained in hESCs cultured in the presence of activin or with inhibition of activin–NODAL signalling for 2 h with SB431542 (n = 3 cultures). Note that the sum of the percentages within each graph is not 100% because some m6A peaks overlap several feature types. MetDiff is an exome peak caller, and, accordingly, 100% of peaks map to exons. MACS2 identifies peaks throughout the genome. d, Venn diagrams showing the overlap of peaks identified by the two pipelines. Only MetDiff peaks that were also identified by MACS2 were considered for subsequent analyses focused on m6A peaks on exons. e, Top sequence motifs identified de novo on all m6A exon peaks, or on those that showed significant downregulation following inhibition of activin–NODAL signalling (activin–NODAL-sensitive m6A peaks; Supplementary Table 2). The position of the methylated adenosine is indicated by a box. f, Coverage profiles for all m6A exon peaks across the length of different genomic features. Each feature type is expressed as 100 bins of equal length with 5′ to 3′ directionality. g, h, Overlap of m6A exon peaks and transcription start sites (TSS) or transcription end sites (TES). In g, the analysis was performed for all m6A peaks. In h, only activin–NODAL-sensitive peaks were considered. i, Left, activin–NODAL-sensitive m6A exon peaks were evaluated for direct overlap with SMAD2/3-binding sites as indicated by ChIP–seq30. n = 482 peaks; FDR = 0.41 as calculated by the permutation test implemented by the GAT python package; N.S., not significant based on 95% confidence interval. Right, overlap was calculated after the same features were mapped to their corresponding transcripts or genes, respectively. n = 372 genes; hypergeometric test P = 2.88 ×10−18, significant based on 95% confidence interval. j, m6A NeMeRIP–seq results for selected transcripts (n = 3 cultures; replicates combined for visualization). Coverage tracks represent read enrichments normalized by million mapped reads and size of the library. Blue, sequencing results of m6A NeMeRIP; orange, sequencing results of pre-NeMeRIP input RNA (negative control). GENCODE gene annotations are shown (red, protein coding exons; white, untranslated exons; all potential exons are shown and overlaid). The location of SMAD2/3 ChIP–seq-binding sites is also shown. Compared to the other genes shown, m6A levels on SOX2 were unaffected by inhibition of activin–NODAL signalling, showing specificity of action. POU5F1 (also known as OCT4) is used as a negative control since it is known to not have a m6A site23, as confirmed by the lack of m6A enrichment compared to the input.
Extended Data Figure 6 Features of activin–NODAL-sensitive differential m6A deposition.
a, Scatter plot of the average log2 fold-change in SB431542 versus activin-treated hESCs for m6A NeMeRIP–seq and pre-NeMeRIP input RNA (n = 3 cultures). The analysis was performed for all m6A exon peaks (left), or for those peaks that were significantly downregulated following inhibition of activin–NODAL signalling (right). Data are colour coded according to the square of the difference between the two values (square diff.). b, c, As Extended Data Fig. 5j, but for representative transcripts that are stably expressed following inhibition of activin–NODAL signalling for 2 h (n = 3 cultures; replicates combined for visualization). The m6A NeMeRIP and input tracks were separated and are shown at different scales to facilitate comparison between the conditions. The m6A peaks and those significantly downregulated after 2 h of SB431542 treatment are indicated. d, Venn diagram illustrating the strategy for identification of m6A peaks on introns. Peaks mapping to the transcribed genome were obtained by running MetDiff using an extended transcriptome annotation based on the pre-NeMeRIP input RNA, which has a high abundance of introns. The resulting peaks were first filtered by overlap with genome-wide MACS2-identified peaks, and then by lack of overlap with annotated exons. e, Results of MetDiff differential methylation analysis in activin versus 2 h SB431542 treatment for m6A peaks on introns. n = 3 cultures; P value calculated by likelihood ratio test implemented in the MetDiff R package, and adjusted to FDR by Benjamini–Hochberg correction. See Supplementary Table 2 for the FDR of individual peaks. Abs.FC, absolute fold-change. f, As Extended Data Fig. 5j, but for a representative transcript that shows activin–NODAL-sensitive m6A deposition in introns (n = 3 cultures; replicates combined for visualization). The m6A peaks on exons, introns, and those significantly downregulated after SB431542 treatment within each subset are indicated. g, Plots of RPKM-normalized mean m6A coverage for m6A exon peaks significantly downregulated after SB431542 treatment (absolute fold-change > 1.5). Data for all such peaks is in blue, whereas green lines report coverage for only those peaks characterized by next generation sequencing reads that span exon–intron junctions. Exons were scaled proportionally, and the positions of the 3′ and 5′ splice sites (SS) are indicated. A window of 500 bp on either side of the splice sites is shown. m6A, signal from m6A NeMeRIP–seq; input, signal from pre-NeMeRIP input RNA. The results show that coverage of activin–NODAL-sensitive m6A peaks often spans across splice sites (highlighted by the dotted lines). h, Heat map representing in an extended form the data shown in g for all activin–NODAL-sensitive m6A exon peaks in hESCs cultured in the presence of activin. There are multiple regions in which sequencing coverage extends across exon–intron junctions (see Supplementary Table 2). i, Example of an activin–NODAL-sensitive peak located in the proximity of a 3′ splice site (n = 3 cultures; replicates combined for visualization). This peak is shown within its genomic context in c, where it is indicated by a dotted box. Top, m6A NeMeRIP–seq coverage; bottom, individual next generation sequencing reads. Multiple reads span the exon–intron junction (indicated by the dashed line). j, Relationship between the decrease of m6A on the most affected exonic peak located on a transcript (y axis) and the mean change of all other peaks mapping to the same transcript (x axis). The analysis considered transcripts with multiple m6A peaks and with at least one peak significantly decreasing after inhibition of activin–NODAL signalling with SB431542 (absolute fold-change > 1.5). Sensitivity of m6A deposition to activin–NODAL signalling across these transcripts is correlated.
Extended Data Figure 7 Generation and functional characterization of hPSCs following iKD of the subunits of the m6A methyltransferase complex.
a, qPCR validation of iKD hESCs cultured in the presence of tetracycline for five days (TET) to drive gene knockdown. Two distinct shRNAs and multiple clonal sublines (cl) were tested for each gene. Expression is normalized to the mean level in hESCs carrying a negative control scrambled (SCR) shRNA. For each gene, sh1 cl1 was selected for further analyses. The mean is indicated, n = 2 cultures. b, Western blot validation of selected iKD hESCs for the indicated genes. TUB4A4 (α-tubulin), loading control. Results are representative of three independent experiments. c, MeRIP–qPCR in iKD hESCs cultured for ten days in the absence (CTR) or presence of tetracycline (TET). m6A abundance is shown relative to control conditions in the same hESC line. The mean is shown, n = 2 technical replicates. Results are representative of two independent experiments. d, m6A dot blot in WTAP or scramble shRNA control iKD hESCs treated as described in c. Decreasing amounts of mRNA were spotted to allow semiquantitative comparisons, as indicated. Results are representative of two independent experiments. e, Immunofluorescence of the pluripotency markers NANOG and OCT4 in iKD hESCs cultured for three passages (15 days) in the absence (CTR) or presence of tetracycline (TET). Scale bars, 100 μm. Results are representative of two independent experiments. f, Flow cytometry showing NANOG expression in cells treated as in e. The percentage and median fluorescence intensity (MFI) of NANOG-positive cells (NANOG+) are shown. The gates used for the analysis are indicated, and were determined on the basis of a secondary-antibody-only negative staining (NEG). Results are representative of two independent experiments.
Extended Data Figure 8 Function of the m6A methyltransferase complex during germ-layer specification.
a, qPCR analysis following neuroectoderm or endoderm differentiation of iKD hESCs cultured in absence (CTR) or presence of tetracycline (TET). Tetracycline treatment was initiated in undifferentiated hESCs for ten days and was maintained during differentiation (three days). Expression was normalized against the mean level in undifferentiated hESCs. Mean ± s.e.m., n = 3 cultures. Significant differences versus the same iKD line in control conditions were calculated by two-way ANOVA with post hoc Holm–Sidak comparisons. b, Flow cytometry quantification of the percentage of SOX1-positive cells (SOX1+) in cells treated as in a. Mean is shown, n = 2 cultures. c, Immunofluorescence of the lineage marker SOX17 in endoderm-differentiated hESCs treated as in a. Scale bars, 100μm. Results are representative of two independent experiments. d, qPCR validation of multiple inducible knockdown (MiKD) hESCs simultaneously expressing shRNAs against WTAP, METTL3 and METTL14. Cells expressing three copies of the scrambled shRNA (SCR3×) were used as negative control. Cells were cultured in the presence of tetracycline (TET) for five days to drive gene knockdown. Mean ± s.e.m., n = 3 cultures. Significant differences versus SCR3× hESCs in control conditions were calculated by two-way ANOVA with post hoc Holm–Sidak comparisons. e, f, qPCR analysis following endoderm differentiation of WTAP, METTL3 and METTL14-MiKD hESCs treated as described in a. Mean ± s.e.m., n = 3 cultures. Significant differences versus control conditions were calculated by two tailed t-test (e) or two-way ANOVA with post hoc Holm–Sidak comparisons (f).
Extended Data Figure 9 Function of the m6A methyltransferase complex during exit from pluripotency induced by inhibition of activin–NODAL signalling.
a, qPCR analyses in iKD hESCs cultured in absence (CTR) or presence (TET) of tetracycline for ten days, then subjected to inhibition of activin–NODAL signalling with SB431542 (SB) for the indicated time (see Extended Data Fig. 10a). Activin, cells maintained in standard pluripotency-promoting culture conditions containing activin and collected at the beginning of the experiment. Mean ± s.e.m., n = 3 cultures. Significant differences versus same iKD line in control conditions were calculated by two-way ANOVA with post hoc Holm–Sidak comparisons. b, Western blots of cells treated as described in a. TUBA4A, loading control. Results are representative of two independent experiments. c, Measurement of mRNA stability in WTAP iKD hESCs cultured in absence (CTR) or presence (TET) of tetracycline for ten days. Samples were collected following transcriptional inhibition using actinomycin D (ActD) for the indicated time. The statistical significance of differences between the mRNA half lives in tetracycline versus control is shown (n = 3 cultures, comparison of fits to one-phase decay model by extra sum-of-squares F-test). The difference was significant for NANOG but not for SOX2 (95% confidence interval). d, Model showing the interplays between activin–NODAL signalling and m6A deposition in hPSCs (left), and the phenotype induced by impairment of the m6A methyltransferase complex (right).
Extended Data Figure 10 Genome-wide analysis of the relationship between WTAP and activin–NODAL signalling.
a, Schematic of the experimental approach to investigate the transcriptional changes induced by the knockdown of the m6A methyltransferase complex subunits during neuroectoderm specification of hESCs. b, qPCR analyses of WTAP iKD hESCs subjected to the experiment outlined in a (n = 3 cultures). Activin, cells maintained in standard pluripotency-promoting culture conditions containing activin and collected at the beginning of the experiment. Z-scores indicate differential expression measured in number of standard deviations from the mean across all time points. c, RNA-seq analysis at selected time points from the samples shown in panel b (n = 3 cultures). The heat map shows Z-scores for the top 5% differentially expressed genes (1789 genes as ranked by the Hotelling T2 statistic). Genes and samples were clustered based on their Euclidean distance, and the four major gene clusters are indicated (see Supplementary Discussion). d, Expression profiles of genes belonging to the clusters indicated in c. Selected results of gene-enrichment analysis and representative genes for each cluster are shown (cluster 1: n = 456 genes; cluster 2: n = 471 genes; cluster 3: n = 442 genes; cluster 4: n = 392 genes; Fisher’s exact test followed by Benjamini–Hochberg correction for multiple comparisons). e, Principal component analysis of RNA-seq results in c (n = 3 cultures). The top 5% differentially expressed genes were considered for this analysis. For each of the two main principal components (PC1 and PC2), the fraction of inter-sample variance that they explain and their proposed biological meaning are reported. f, Proportion of transcripts marked by at least one high-confidence m6A peak23 in transcripts significantly up- or downregulated following WTAP iKD in hESCs maintained in the presence of activin (left), or following inhibition of activin–NODAL signalling for 2 h with SB431542 in control cells (right). Differential gene expression was calculated in three cultures using the negative binomial test implemented in DEseq2 with a cutoff of P < 0.05 and abs.FC > 2. The number of genes in each group and the hypergeometric probabilities of the observed overlaps with m6A-marked transcripts are reported (n.s.: not significant at 95% confidence interval).
Supplementary information
Supplementary Information
This file contains a Supplementary Discussion and Supplementary References. (PDF 244 kb)
Supplementary Figure 1
This file contains uncropped western blot scans with size marker indications. (PDF 2049 kb)
Supplementary Table 1
This file contains SMAD2/3 interacting proteins in hESCs and endoderm cells. (XLSX 23 kb)
Supplementary Table 2
This file contains m6A peaks in exons, introns, and spanning exon-intron junctions. (XLSX 1652 kb)
Supplementary Table 3
This file contains gene ontology enrichment analysis for transcripts where m6A is downregulated after inhibition of Activin/Nodal signalling. (XLSX 37 kb)
Supplementary Table 4
This file contains genes differentially expressed during exit from pluripotency in WTAP inducible knockdown hESCs. (XLSX 405 kb)
Supplementary Table 5
This file contains a list of oligonucleotides used in the study. (XLSX 19 kb)
Supplementary Table 6
This file contains a list of antibodies used in the study. (XLSX 39 kb)
Source data
Rights and permissions
About this article
Cite this article
Bertero, A., Brown, S., Madrigal, P. et al. The SMAD2/3 interactome reveals that TGFβ controls m6A mRNA methylation in pluripotency. Nature 555, 256–259 (2018). https://doi.org/10.1038/nature25784
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature25784
This article is cited by
-
The role of the methyltransferase METTL3 in prostate cancer: a potential therapeutic target
BMC Cancer (2024)
-
Fibroblast-specific PRMT5 deficiency suppresses cardiac fibrosis and left ventricular dysfunction in male mice
Nature Communications (2024)
-
Bioinformatic Analysis of m6A Regulator-Mediated RNA Methylation Modification Patterns and Immune Microenvironment Characterization in Endometriosis
Biochemical Genetics (2024)
-
Comprehensive analysis of m6A regulators characterized by the immune microenvironment in Duchenne muscular dystrophy
Journal of Translational Medicine (2023)
-
CCL3 secreted by hepatocytes promotes the metastasis of intrahepatic cholangiocarcinoma by VIRMA-mediated N6-methyladenosine (m6A) modification
Journal of Translational Medicine (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.