Abstract
Dopamine is a neurotransmitter that has been implicated in processes as diverse as reward, addiction, control of coordinated movement, metabolism and hormonal secretion. Correspondingly, dysregulation of the dopaminergic system has been implicated in diseases such as schizophrenia, Parkinson’s disease, depression, attention deficit hyperactivity disorder, and nausea and vomiting. The actions of dopamine are mediated by a family of five G-protein-coupled receptors1. The D2 dopamine receptor (DRD2) is the primary target for both typical2 and atypical3,4 antipsychotic drugs, and for drugs used to treat Parkinson’s disease. Unfortunately, many drugs that target DRD2 cause serious and potentially life-threatening side effects due to promiscuous activities against related receptors4,5. Accordingly, a molecular understanding of the structure and function of DRD2 could provide a template for the design of safer and more effective medications. Here we report the crystal structure of DRD2 in complex with the widely prescribed atypical antipsychotic drug risperidone. The DRD2–risperidone structure reveals an unexpected mode of antipsychotic drug binding to dopamine receptors, and highlights structural determinants that are essential for the actions of risperidone and related drugs at DRD2.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Missale, C., Nash, S. R., Robinson, S. W., Jaber, M. & Caron, M. G. Dopamine receptors: from structure to function. Physiol. Rev. 78, 189–225 (1998)
Creese, I., Burt, D. R. & Snyder, S. H. Dopamine receptor binding predicts clinical and pharmacological potencies of antischizophrenic drugs. Science 192, 481–483 (1976)
Meltzer, H. Y., Matsubara, S. & Lee, J.-C. Classification of typical and atypical antipsychotic drugs on the basis of dopamine D-1, D-2 and serotonin2 pKi values. J. Pharmacol. Exp. Ther. 251, 238–246 (1989)
Roth, B. L., Sheffler, D. J. & Kroeze, W. K. Magic shotguns versus magic bullets: selectively non-selective drugs for mood disorders and schizophrenia. Nat. Rev. Drug Discov. 3, 353–359 (2004)
Roth, B. L. Drugs and valvular heart disease. N. Engl. J. Med. 356, 6–9 (2007)
Seeman, P. & Lee, T. Antipsychotic drugs: direct correlation between clinical potency and presynaptic action on dopamine neurons. Science 188, 1217–1219 (1975)
Sibley, D. R. & Monsma, F. J. Jr. Molecular biology of dopamine receptors. Trends Pharmacol. Sci. 13, 61–69 (1992)
Beaulieu, J. M. & Gainetdinov, R. R. The physiology, signaling, and pharmacology of dopamine receptors. Pharmacol. Rev. 63, 182–217 (2011)
Volkow, N. D., Fowler, J. S., Wang, G. J., Swanson, J. M. & Telang, F. Dopamine in drug abuse and addiction: results of imaging studies and treatment implications. Arch. Neurol. 64, 1575–1579 (2007)
Bunzow, J. R. et al. Cloning and expression of a rat D2 dopamine receptor cDNA. Nature 336, 783–787 (1988)
Grandy, D. K. et al. Cloning of the cDNA and gene for a human D2 dopamine receptor. Proc. Natl Acad. Sci. USA 86, 9762–9766 (1989)
Monsma, F. J., Jr, McVittie, L. D., Gerfen, C. R., Mahan, L. C. & Sibley, D. R. Multiple D2 dopamine receptors produced by alternative RNA splicing. Nature 342, 926–929 (1989)
Allen, J. A. et al. Discovery of β-arrestin-biased dopamine D2 ligands for probing signal transduction pathways essential for antipsychotic efficacy. Proc. Natl Acad. Sci. USA 108, 18488–18493 (2011)
Javitch, J. A., Fu, D., Chen, J. & Karlin, A. Mapping the binding-site crevice of the dopamine D2 receptor by the substituted-cysteine accessibility method. Neuron 14, 825–831 (1995)
Ballesteros, J. A., Shi, L. & Javitch, J. A. Structural mimicry in G protein-coupled receptors: implications of the high-resolution structure of rhodopsin for structure-function analysis of rhodopsin-like receptors. Mol. Pharmacol. 60, 1–19 (2001)
Chien, E. Y. et al. Structure of the human dopamine D3 receptor in complex with a D2/D3 selective antagonist. Science 330, 1091–1095 (2010)
Wang, S. et al. D4 dopamine receptor high-resolution structures enable the discovery of selective agonists. Science 358, 381–386 (2017)
Manglik, A. et al. Structure-based discovery of opioid analgesics with reduced side effects. Nature 537, 185–190 (2016)
Wacker, D., Stevens, R. C. & Roth, B. L. How ligands illuminate GPCR molecular pharmacology. Cell 170, 414–427 (2017)
McCorvy, J. D. et al. Structure-inspired design of β-arrestin-biased ligands for aminergic GPCRs. Nat. Chem. Biol. 14, 126–134 (2018)
Free, R. B. et al. Discovery and characterization of a G protein-biased agonist that inhibits β-arrestin recruitment to the D2 dopamine receptor. Mol. Pharmacol. 86, 96–105 (2014)
Roberts, D. J. & Strange, P. G. Mechanisms of inverse agonist action at D2 dopamine receptors. Br. J. Pharmacol. 145, 34–42 (2005)
Rasmussen, S. G. et al. Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature 477, 549–555 (2011)
Shapiro, D. A., Kristiansen, K., Weiner, D. M., Kroeze, W. K. & Roth, B. L. Evidence for a model of agonist-induced activation of 5–HT2A serotonin receptors which involves the disruption of a strong ionic interaction between helices 3 and 6. J. Biol. Chem. 18, 11441–11449 (2002)
Ballesteros, J. A. et al. Activation of the β2-adrenergic receptor involves disruption of an ionic lock between the cytoplasmic ends of transmembrane segments 3 and 6. J. Biol. Chem. 276, 29171–29177 (2001)
Palczewski, K. et al. Crystal structure of rhodopsin: a G protein-coupled receptor. Science 289, 739–745 (2000)
Janssen, P. A. et al. Pharmacology of risperidone (R 64 766), a new antipsychotic with serotonin–S2 and dopamine–D2 antagonistic properties. J. Pharmacol. Exp. Ther. 244, 685–693 (1988)
Kapur, S., Zipursky, R., Jones, C., Remington, G. & Houle, S. Relationship between dopamine D2 occupancy, clinical response, and side effects: a double-blind PET study of first-episode schizophrenia. Am. J. Psychiatry 157, 514–520 (2000)
Kapur, S. & Seeman, P. Does fast dissociation from the dopamine D2 receptor explain the action of atypical antipsychotics?: A new hypothesis. Am. J. Psychiatry 158, 360–369 (2001)
Sykes, D. A. et al. Extrapyramidal side effects of antipsychotics are linked to their association kinetics at dopamine D2 receptors. Nat. Commun. 8, 763 (2017)
Rosenbaum, D. M. et al. GPCR engineering yields high-resolution structural insights into β2-adrenergic receptor function. Science 318, 1266–1273 (2007)
Caffrey, M. & Cherezov, V. Crystallizing membrane proteins using lipidic mesophases. Nat. Protocols 4, 706–731 (2009)
Minor, W., Cymborowski, M., Otwinowski, Z. & Chruszcz, M. HKL-3000: the integration of data reduction and structure solution—from diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 62, 859–866 (2006)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010)
Motulsky, H. J. & Mahan, L. C. The kinetics of competitive radioligand binding predicted by the law of mass action. Mol. Pharmacol. 25, 1–9 (1984)
Pei, J. & Grishin, N. V. PROMALS3D: multiple protein sequence alignment enhanced with evolutionary and three-dimensional structural information. Methods Mol. Biol. 1079, 263–271 (2014)
Webb, B. & Sali, A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformatics 47, 5 6 1–5 6 32 (2014)
Coleman, R. G., Carchia, M., Sterling, T., Irwin, J. J. & Shoichet, B. K. Ligand pose and orientational sampling in molecular docking. PLoS One 8, e75992 (2013)
Southan, C. et al. The IUPHAR/BPS Guide to pharmacology in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. Nucleic Acids Res. 44 (D1), D1054–D1068 (2016)
Mysinger, M. M., Carchia, M., Irwin, J. J. & Shoichet, B. K. Directory of useful decoys, enhanced (DUD-E): better ligands and decoys for better benchmarking. J. Med. Chem. 55, 6582–6594 (2012)
Case, D. A . et al. AMBER 2015. (University of California, 2015)
Word, J. M., Lovell, S. C., Richardson, J. S. & Richardson, D. C. Asparagine and glutamine: using hydrogen atom contacts in the choice of side-chain amide orientation. J. Mol. Biol. 285, 1735–1747 (1999)
Gallagher, K. & Sharp, K. Electrostatic contributions to heat capacity changes of DNA-ligand binding. Biophys. J. 75, 769–776 (1998)
Sharp, K. A. Polyelectrolyte electrostatics: Salt dependence, entropic, and enthalpic contributions to free energy in the nonlinear Poisson–Boltzmann model. Biopolymers 36, 227–243 (1995)
Mysinger, M. M. & Shoichet, B. K. Rapid context-dependent ligand desolvation in molecular docking. J. Chem. Inf. Model. 50, 1561–1573 (2010)
Sadowski, J., Gasteiger, J. & Klebe, G. Comparison of automatic three-dimensional model builders using 639 X-ray structures. J. Chem. Inf. Comput. Sci. 34, 1000–1008 (1994)
Hawkins, P. C., Skillman, A. G., Warren, G. L., Ellingson, B. A. & Stahl, M. T. Conformer generation with OMEGA: algorithm and validation using high quality structures from the Protein Databank and Cambridge Structural Database. J. Chem. Inf. Model. 50, 572–584 (2010)
Chambers, C. C., Hawkins, G. D., Cramer, C. J. & Truhlar, D. G. Model for aqueous solvation based on class IV atomic charges and first solvation shell effects. J. Phys. Chem. 100, 16385–16398 (1996)
Li, J., Zhu, T., Cramer, C. J. & Truhlar, D. G. New class IV charge model for extracting accurate partial charges from wave functions. J. Phys. Chem. A 102, 1820–1831 (1998)
Acknowledgements
This work was supported by NIH Grants RO1MH61887, U19MH82441, the NIMH Psychoactive Drug Screening Program Contract and the Michael Hooker Chair for Protein Therapeutics and Translational Proteomics (to B.L.R.) and by R35GM122481 (to B.K.S.). We thank J. Sondek and S. Endo-Streeter for providing independent structure quality control analysis; M. J. Miley and the UNC macromolecular crystallization core for advice and use of their equipment for crystal harvesting and transport, which is supported by the National Cancer Institute under award number P30CA016086; B. E. Krumm for advice on data processing and help with thermostabilization assays; and the staff of GM/CA@APS, which has been funded with Federal funds from the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006). This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science user facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
Author information
Authors and Affiliations
Contributions
S.W. designed experiments, developed the DRD2 construct and purification, expressed, purified and crystallized the receptor, collected diffraction data, solved and refined the structure, analysed the structure, performed radioligand binding and prepared the manuscript. T.C. performed radioligand binding, analysed the data and assisted with preparing the manuscript. A.L. conducted the homology modelling and docking and helped to edit the manuscript. B.K.S. supervised the modelling and docking and helped to prepare the manuscript. D.W. refined and analysed the structure, supervised the structure determination and assisted with preparing the manuscript. B.L.R. supervised the overall project and management and prepared the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks D. Sibley and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 Thermostability of DRD2 constructs, crystal packing of the DRD2–risperidone complex and representative electron density of the DRD2 structure.
a, Membranes containing DRD2 or DRD2 with thermostability mutations were heated for 30 min with 1 nM [3H]-N-methylspiperone and the amount of bound [3H]-ligand was determined. b, Purified DRD2–T4L protein (with or without thermostability mutations) was heated with 10 μM risperidone and 1 μM BODIPY FL l-cystine dye using a temperature gradient and the amount of dye bound to unfolding protein was determined. Data were analysed by nonlinear regression and apparent Tm values (transition temperature where 50% of the receptor is inactive) were determined from analysis of the sigmoidal melting curves. All data in a and b are mean ± s.e.m. of three independent assays. c–e, Packing of the DRD2–risperidone complex crystallized in the P212121 spacegroup. DRD2 is shown in green and the T4L-fusion protein is shown in red, or in cyan where it interacts with DRD2. EL1 and EL2 of DRD2 are shown in magenta and blue, respectively. f, 2Fo–Fc electron density map (blue mesh) of risperidone (yellow) contoured at 1σ. g, Fo–Fc omit map (green mesh) contoured at 3.0σ of risperidone (yellow). h, 2 Fo–Fc electron density map of DRD2 binding pocket residues (blue mesh) contoured at 1σ.
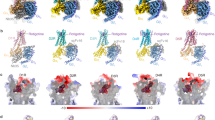
Extended Data Figure 2 Conserved hydrophobic residue of EL2 in all available aminergic receptor structures.
In all panels, receptors are shown as cartoons. Ligands and residues are shown as sticks. a, 5HT1B (PDB code: 4IAR). b, 5HT2B (PDB code: 5TVN). c, DRD2. d, DRD3 (PDB code: 3PBL). e, DRD4 (PDB code: 5WIU). f, ACM1 (PDB code: 5CXV). g, ACM2 (PDB code: 3UON). h, ACM3 (PDB code: 4ADJ). i, ACM4 (PDB code: 4DSG). j, HRH1 (PDB code: 3RZE). k, ADRB1 (PDB code: 2VT4). l, ADRB2 (PDB code: 2RH1). m, DRD2. n, Conserved EL2 hydrophobic residues (red box) are located two residues away from a conserved cysteine that forms a disulphide bridge between EL2 and TMIII. Notable exceptions to the presence of a hydrophobic residue are DRD1 and DRD5, which contain a serine, and HRH1 and HRH4, which contain a threonine and proline, respectively.
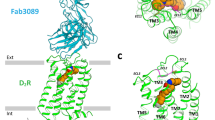
Extended Data Figure 3 Comparison of D2 receptors viewed from the extracellular side, and structural alignment with β2AR and A2AR reveals an inactive state of DRD2.
a–d, DRD2, green; DRD3, magenta (PDB code: 3PBL); DRD4, blue (PDB code: 5WIU). Risperidone (yellow), eticlopride (cyan) and nemonapride (light pink) are shown as sticks and spheres. Displacements of H6.55 and Y/V7.35 are shown at DRD2 (a), DRD3 (b) and DRD4 (c). d, Views from the extracellular side of DRD2 and DRD3. e, f, Superposition of TMVI at DRD2 (green), inactive β2AR (yellow, PDB code: 2RH1), active β2AR (light pink, PDB code: 3SN6), inactive A2AR (brown, PDB code: 3REY) and active A2AR (blue, PDB code: 5G53) aligned through helices I–IV. g–j, Cytoplasmic view of alignment between DRD2 and active and inactive β2AR (g, h) or A2AR (i, j). Rearrangements of two highly conserved residues (Y7.53 and R3.50) within the core of the receptor are shown as sticks. Ligands are omitted for clarity and hydrogen bonds are shown as grey dotted lines.
Extended Data Figure 4 Conserved Trp of EL1 in all available aminergic receptor structures shows its unique position in DRD2–risperidone.
Receptors are shown as cartoons. Ligands and residues are shown as sticks. a, Conserved Trp residues of EL1 are shown in red boxes. b, 5HT1B (PDB code: 4IAR). c, 5HT2B (PDB code: 5TVN). d, DRD2. e, DRD3 (PDB code: 3PBL). f, DRD4 (PDB code: 5WIU). g, ACM1 (PDB code: 5CXV). h, ACM2 (PDB code: 3UON). i, ACM3 (PDB code: 4ADJ). j, ACM4 (PDB code: 4DSG). k, HRH1 (PDB code: 3RZE). l, ADRB1 (PDB code: 2VT4). m, ADRB2 (PDB code: 2RH1).
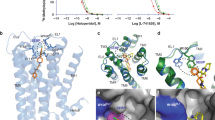
Extended Data Figure 5 Risperidone has distinct poses in solution and in complex with DRD2, and comparison of X-ray structure and model of DRD2.
a, Trp100EL1 determines the configuration of the tetrahydropyridopyrimidinone moiety of risperidone. Structure of unbound risperidone shown in green and DRD2-bound risperidone shown in yellow. b, Electron density (2Fo–Fc maps, blue mesh) for W100EL1 in the DRD2–risperidone complex (contoured at 1.0σ). c, 2Fo–Fc electron density map (blue mesh) of Leu942.64, Trp100EL1, Ile184EL2 and risperidone (yellow) contoured at 0.8σ. d, Overall view of DRD2–risperidone X-ray structure and model. e–h, Comparison of X-ray structure and model of DRD2. In d–h, DRD2 X-ray structure and model are shown as cartoons, with the X-ray structure in green and the model in magenta or blue. Risperidone is shown in the X-ray structure as yellow spheres or sticks, and in the model as cyan or light pink.
Extended Data Figure 6 Patch residues of the DRD2 orthosteric pocket impair the dissociation rates of risperidone, aripiprazole, N-methylspiperone and nemonapride.
a–g, Comparison of risperidone dissociation from wild-type DRD2 (a) and W100EL1A (b), W100EL1L (c), W100EL1F (d), L942.64A (e), I184EL2A (f) or L942.64A/I184EL2A (g) mutants. h–n, Comparison of aripiprazole dissociation from wild-type DRD2 (h) and W100EL1A (i), W100EL1L (j), W100EL1F (k), L942.64A (l), I184EL2A (m) or L942.64A/I184EL2A (n) mutants. o, p, Comparison of N-methylspiperone (o) or nemonapride (p) dissociation from wild-type DRD2 and W100EL1A, W100EL1L or W100EL1F mutants (n = 3). q, r, Comparison of N-methylspiperone (q) or nemonapride (r) dissociation from wild-type DRD2 and L942.64A, I184EL2A or L942.64A/I184EL2A mutants. All data are mean ± s.e.m. of four independent assays (n = 4 independent experiments). Error bars in o–r denote s.e.m. from four independent assays.
Supplementary information
Rights and permissions
About this article
Cite this article
Wang, S., Che, T., Levit, A. et al. Structure of the D2 dopamine receptor bound to the atypical antipsychotic drug risperidone. Nature 555, 269–273 (2018). https://doi.org/10.1038/nature25758
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature25758
This article is cited by
-
Antiemetic activity of abietic acid possibly through the 5HT3 and muscarinic receptors interaction pathways
Scientific Reports (2024)
-
Differential Effects of Neonatal Ventral Hippocampus Lesion on Behavior and Corticolimbic Plasticity in Wistar–Kyoto and Spontaneously Hypertensive Rats
Neurochemical Research (2024)
-
Sequestration of Gβγ by deubiquitinated arrestins into the nucleus as a novel desensitization mechanism of G protein–coupled receptors
Cell Communication and Signaling (2023)
-
Structure, function and drug discovery of GPCR signaling
Molecular Biomedicine (2023)
-
Structural basis of amine odorant perception by a mammal olfactory receptor
Nature (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.