Abstract
N-glycosylation is a ubiquitous modification of eukaryotic secretory and membrane-bound proteins; about 90% of glycoproteins are N-glycosylated. The reaction is catalysed by an eight-protein oligosaccharyltransferase (OST) complex that is embedded in the endoplasmic reticulum membrane. Our understanding of eukaryotic protein N-glycosylation has been limited owing to the lack of high-resolution structures. Here we report a 3.5 Å resolution cryo-electron microscopy structure of the Saccharomyces cerevisiae OST complex, revealing the structures of subunits Ost1–Ost5, Stt3, Wbp1 and Swp1. We found that seven phospholipids mediate many of the inter-subunit interactions, and an Stt3 N-glycan mediates interactions with Wbp1 and Swp1 in the lumen. Ost3 was found to mediate the OST–Sec61 translocon interface, funnelling the acceptor peptide towards the OST catalytic site as the nascent peptide emerges from the translocon. The structure provides insights into co-translational protein N-glycosylation, and may facilitate the development of small-molecule inhibitors that target this process.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Larkin, A. & Imperiali, B. The expanding horizons of asparagine-linked glycosylation. Biochemistry 50, 4411–4426 (2011)
Cherepanova, N., Shrimal, S. & Gilmore, R. N-linked glycosylation and homeostasis of the endoplasmic reticulum. Curr. Opin. Cell Biol. 41, 57–65 (2016)
Dempski, R. E. Jr & Imperiali, B. Oligosaccharyl transferase: gatekeeper to the secretory pathway. Curr. Opin. Chem. Biol. 6, 844–850 (2002)
Mohorko, E., Glockshuber, R. & Aebi, M. Oligosaccharyltransferase: the central enzyme of N-linked protein glycosylation. J. Inherit. Metab. Dis. 34, 869–878 (2011)
Kornfeld, R. & Kornfeld, S. Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 54, 631–664 (1985)
Helenius, A. & Aebi, M. Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 73, 1019–1049 (2004)
Apweiler, R., Hermjakob, H. & Sharon, N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim. Biophys. Acta 1473, 4–8 (1999)
Hennet, T. & Cabalzar, J. Congenital disorders of glycosylation: a concise chart of glycocalyx dysfunction. Trends Biochem. Sci. 40, 377–384 (2015)
Nothaft, H. & Szymanski, C. M. Protein glycosylation in bacteria: sweeter than ever. Nat. Rev. Microbiol. 8, 765–778 (2010)
Matsumoto, S., Taguchi, Y., Shimada, A., Igura, M. & Kohda, D. Tethering an N-glycosylation sequon-containing peptide creates a catalytically competent oligosaccharyltransferase complex. Biochemistry 56, 602–611 (2017)
Matsumoto, S. et al. Crystal structures of an archaeal oligosaccharyltransferase provide insights into the catalytic cycle of N-linked protein glycosylation. Proc. Natl Acad. Sci. USA 110, 17868–17873 (2013)
Napiórkowska, M. et al. Molecular basis of lipid-linked oligosaccharide recognition and processing by bacterial oligosaccharyltransferase. Nat. Struct. Mol. Biol. 24, 1100–1106 (2017)
Lizak, C., Gerber, S., Numao, S., Aebi, M. & Locher, K. P. X-ray structure of a bacterial oligosaccharyltransferase. Nature 474, 350–355 (2011)
Kelleher, D. J. & Gilmore, R. An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology 16, 47R–62R (2006)
Yan, A. & Lennarz, W. J. Two oligosaccharyl transferase complexes exist in yeast and associate with two different translocons. Glycobiology 15, 1407–1415 (2005)
Shrimal, S., Cherepanova, N. A. & Gilmore, R. Cotranslational and posttranslocational N-glycosylation of proteins in the endoplasmic reticulum. Semin. Cell Dev. Biol. 41, 71–78 (2015)
Schulz, B. L. et al. Oxidoreductase activity of oligosaccharyltransferase subunits Ost3p and Ost6p defines site-specific glycosylation efficiency. Proc. Natl Acad. Sci. USA 106, 11061–11066 (2009)
Mohorko, E. et al. Structural basis of substrate specificity of human oligosaccharyl transferase subunit N33/Tusc3 and its role in regulating protein N-glycosylation. Structure 22, 590–601 (2014)
Gayen, S. & Kang, C. Solution structure of a human minimembrane protein Ost4, a subunit of the oligosaccharyltransferase complex. Biochem. Biophys. Res. Commun. 409, 572–576 (2011)
Zubkov, S., Lennarz, W. J. & Mohanty, S. Structural basis for the function of a minimembrane protein subunit of yeast oligosaccharyltransferase. Proc. Natl Acad. Sci. USA 101, 3821–3826 (2004)
Pathak, R., Hendrickson, T. L. & Imperiali, B. Sulfhydryl modification of the yeast Wbp1p inhibits oligosaccharyl transferase activity. Biochemistry 34, 4179–4185 (1995)
Yan, Q., Prestwich, G. D. & Lennarz, W. J. The Ost1p subunit of yeast oligosaccharyl transferase recognizes the peptide glycosylation site sequence, -Asn-X-Ser/Thr-. J. Biol. Chem. 274, 5021–5025 (1999)
Li, H., Chavan, M., Schindelin, H., Lennarz, W. J. & Li, H. Structure of the oligosaccharyl transferase complex at 12 Å resolution. Structure 16, 432–440 (2008)
Pfeffer, S. et al. Structure of the native Sec61 protein-conducting channel. Nat. Commun. 6, 8403 (2015)
Pfeffer, S. et al. Structure of the mammalian oligosaccharyl-transferase complex in the native ER protein translocon. Nat. Commun. 5, 3072 (2014)
Pfeffer, S. et al. Dissecting the molecular organization of the translocon-associated protein complex. Nat. Commun. 8, 14516 (2017)
Lara, P. et al. Refined topology model of the STT3/Stt3 protein subunit of the oligosaccharyltransferase complex. J. Biol. Chem. 292, 11349–11360 (2017)
Igura, M. et al. Structure-guided identification of a new catalytic motif of oligosaccharyltransferase. EMBO J. 27, 234–243 (2008)
Huang, C., Bhaskaran, R. & Mohanty, S. Eukaryotic N-glycosylation occurs via the membrane-anchored C-terminal domain of the Stt3p subunit of oligosaccharyltransferase. J. Biol. Chem. 287, 32450–32458 (2012)
Kowarik, M. et al. Definition of the bacterial N-glycosylation site consensus sequence. EMBO J. 25, 1957–1966 (2006)
Shrimal, S., Trueman, S. F. & Gilmore, R. Extreme C-terminal sites are posttranslocationally glycosylated by the STT3B isoform of the OST. J. Cell Biol. 201, 81–95 (2013)
Li, G., Yan, Q., Nita-Lazar, A., Haltiwanger, R. S. & Lennarz, W. J. Studies on the N-glycosylation of the subunits of oligosaccharyl transferase in Saccharomyces cerevisiae. J. Biol. Chem. 280, 1864–1871 (2005)
Yan, A., Ahmed, E., Yan, Q. & Lennarz, W. J. New findings on interactions among the yeast oligosaccharyl transferase subunits using a chemical cross-linker. J. Biol. Chem. 278, 33078–33087 (2003)
Palsdottir, H. & Hunte, C. Lipids in membrane protein structures. Biochim. Biophys. Acta 1666, 2–18 (2004)
Hunte, C. & Richers, S. Lipids and membrane protein structures. Curr. Opin. Struct. Biol. 18, 406–411 (2008)
Govaerts, C. Lipids can make them stick together. Trends Biochem. Sci. 42, 329–330 (2017)
Bai, X. C. et al. An atomic structure of human γ-secretase. Nature 525, 212–217 (2015)
Beatson, S. & Ponting, C. P. GIFT domains: linking eukaryotic intraflagellar transport and glycosylation to bacterial gliding. Trends Biochem. Sci. 29, 396–399 (2004)
Bause, E., Wesemann, M., Bartoschek, A. & Breuer, W. Epoxyethylglycyl peptides as inhibitors of oligosaccharyltransferase: double-labelling of the active site. Biochem. J. 322, 95–102 (1997)
Chavan, M., Rekowicz, M. & Lennarz, W. Insight into functional aspects of Stt3p, a subunit of the oligosaccharyl transferase. Evidence for interaction of the N-terminal domain of Stt3p with the protein kinase C cascade. J. Biol. Chem. 278, 51441–51447 (2003)
Shrimal, S., Cherepanova, N. A. & Gilmore, R. One flexible loop in OST lassos both substrates. Nat. Struct. Mol. Biol. 24, 1009–1010 (2017)
Kern, N. R. et al. Lipid-linked oligosaccharides in membranes sample conformations that facilitate binding to oligosaccharyltransferase. Biophys. J. 107, 1885–1895 (2014)
Harada, Y., Li, H., Li, H. & Lennarz, W. J. Oligosaccharyltransferase directly binds to ribosome at a location near the translocon-binding site. Proc. Natl Acad. Sci. USA 106, 6945–6949 (2009)
Shrimal, S., Cherepanova, N. A. & Gilmore, R. DC2 and KCP2 mediate the interaction between the oligosaccharyltransferase and the ER translocon. J. Cell Biol. 216, 3625–3638 (2017)
Hakomori, S. Glycosylation defining cancer malignancy: new wine in an old bottle. Proc. Natl Acad. Sci. USA 99, 10231–10233 (2002)
Dube, D. H. & Bertozzi, C. R. Glycans in cancer and inflammation--potential for therapeutics and diagnostics. Nat. Rev. Drug Discov. 4, 477–488 (2005)
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017)
Zhang, K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016)
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Amunts, A. et al. Structure of the yeast mitochondrial large ribosomal subunit. Science 343, 1485–1489 (2014)
Chen, V. B . et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010).
Acknowledgements
Cryo-EM images were collected in the David Van Andel Advanced Cryo-Electron Microscopy Suite at Van Andel Research Institute. We thank Y. Harada for advice on yeast genetics and D. Nadziejka for proofreading. This work was partially supported by Van Andel Research Institute (to H.L.) and the US National Institutes of Health (GM111742 to H.L.).
Author information
Authors and Affiliations
Contributions
L.B. and H.L. designed the project. L.B. and A.K. purified proteins. L.B., T.W. and G.Z. collected cryo-EM data. L.B. processed data. L.B. and H.L. analysed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks S. Withers and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 Identification of Ost3 and Ost6 by mass spectrometry.
a, The Coomassie blue–stained SDS–PAGE gel of the purified OST complex. The small subunits Ost2, Ost4–Flag and Ost5 were not visible in this 12% acrylamide SDS–PAGE gel because of their weak density. b, Sequence coverage of tryptic digestion mass spectrometry of three bands at around 30 kDa that are labelled as Ost3, Ost6 and Swp1. The detected peptides are highlighted in blue. The lower bars under the sequences indicate matched peptides. Darker blue indicates more overlaps of peptides detected. c, Ost2, Ost4–Flag and Ost5 were seen in the 15% acrylamide SDS–PAGE gel that was run slower and stained longer. Experiments in a and c were repeated more than three times with similar results.
Extended Data Figure 2 Single-particle cryo-EM analysis of the OST complex.
a, A representative electron micrograph of the OST complex imaged in the Titan Krios with a K2 detector. About 4,000 similar micrographs were recorded. b, Selected reference-free 2D class averages. c, 2D and 3D image classification procedure. d, Gold-standard Fourier correlation of two independent half maps, and the validation correlation curves of the atomic model by comparing the model with the final map or with the two half maps. e, Local resolution map of the OST complex structure.
Extended Data Figure 3 A gallery of selected regions in the OST structure, illustrating the fitting between the 3D density map and the atomic model.
Selected regions in the structure of the OST complex include 26 TMHs, several regions in the lumenal domains, four selected lipids and two N-glycans.
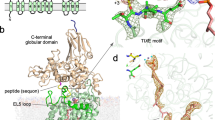
Extended Data Figure 4 Electron density map of the TRX domain of Ost3.
a, b, From 3D classification, one class (class I) contained stronger Ost3 TRX domain density than other classes. This map was further refined to 4.4 Å. Surface view of the map (left) and the corresponding cartoon view of the atomic model (right), coloured by subunit, are shown in two orthogonal side views. The N-terminal TRX domain of Ost3 is highlighted by a magenta disk and is visible in this low-threshold display. The detergent densities that surround the transmembrane region of OST are visible at this threshold, and are coloured in cyan. The structure of the homologous Ost6 TRX (PDB code 3G7Y) is tentatively placed for the purpose of domain location.
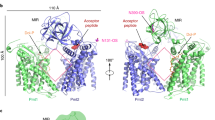
Extended Data Figure 5 The transmembrane region of the OST complex.
a, b, The TMHs of OST form a triangular shape and shown in cytoplasmic view (a) and lumenal view (b). The catalytic subunit Stt3 is in the centre, surrounded by the other subunits. There is a sizable cavity in the centre (red dotted circle). c, Superposition of the transmembrane region of Stt3 and PglB (PDB code 5OGL) viewed from the cytoplasmic side. The Stt3 TMH8–9 (light grey ellipse) moves towards the LLO biding surface relative to the TMH8–9 of PglB (light blue ellipse), creating space for the Ost2 TMHs. The Stt3 TMH1 and TMH13 also move apart, forming a space for the only TMH of Ost4.
Extended Data Figure 6 Sequence alignments of S. cerevisiae Stt3 and A. fulgidus PglB.
PglB does not have the CTE sequence (underscored) found in the yeast Stt3 and human STT3B. Several conserved residues in the active site are highlighted in red. R331 in the PglB, which stabilizes the −2 position D of the acceptor peptide, is highlighted in blue. The asterisk indicates positions with identical residue, a colon indicates strong conservation and a full stop indicates weak conservation.
Extended Data Figure 7 Sequence alignment of selected eukaryotic Stt3.
The CTE of human STT3A is shorter than those of STT3B and yeast Stt3. hs, Homo sapiens; sc, Saccharomyces cerevisiae.
Extended Data Figure 8 Sequence alignment of selected eukaryotic Ost1.
An extra CTD in ribophorin I of complex eukaryotes such as flies, mice and humans is not present in the yeast proteins (shaded grey). NTD1 is shaded in light green, NTD2 in light magenta, TMH in light blue, and the CTD of ribophorin I of complex eukaryotes in light grey. dm, Drosophila melanogaster; pp, Pichia pastoris.
Extended Data Figure 9 Sequence alignment of selected eukaryotic Swp1.
Ribophorin II of complex eukaryotes has evolved an extra N-terminal domain (NTD0, shaded in light orange) in the lumen that is not present in the two yeast proteins.
Supplementary information
Cryo-EM 3D density Map of the S. cerevisiae OST Complex
Surface-rendered cryo-EM 3D map of the OST complex segmented according to the eight individual subunits, which are coloured as in Fig. 1. (MOV 23844 kb)
Atomic Model of the S. cerevisiae OST Complex
Overall structure of the OST complex shown in cartoon. Individual subunits are coloured as in Fig. 1. (MOV 23807 kb)
Atomic Model of the OST-Sec61 Super-Complex
Docking the structures of OST and Sec61 (PDB ID: 3JC2) into the cryo-electron tomogram of a mammalian Ribosome-Sec61-OST-TRAP complex (EMD-3069) reveals the interface between OST and Sec61. (MOV 14448 kb)
Rights and permissions
About this article
Cite this article
Bai, L., Wang, T., Zhao, G. et al. The atomic structure of a eukaryotic oligosaccharyltransferase complex. Nature 555, 328–333 (2018). https://doi.org/10.1038/nature25755
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature25755
This article is cited by
-
Reconstitution and resonance assignments of yeast OST subunit Ost4 and its critical mutant Ost4V23D in liposomes by solid-state NMR
Journal of Biomolecular NMR (2024)
-
Functional classification of DDOST variants of uncertain clinical significance in congenital disorders of glycosylation
Scientific Reports (2023)
-
Inference and reconstruction of the heimdallarchaeial ancestry of eukaryotes
Nature (2023)
-
Molecular basis for glycan recognition and reaction priming of eukaryotic oligosaccharyltransferase
Nature Communications (2022)
-
The structure of an archaeal oligosaccharyltransferase provides insight into the strict exclusion of proline from the N-glycosylation sequon
Communications Biology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.