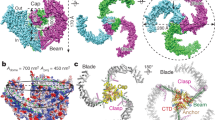
Abstract
Piezo1 and Piezo2 are mechanically activated ion channels that mediate touch perception, proprioception and vascular development. Piezo proteins are distinct from other ion channels and their structure remains poorly defined, which impedes detailed study of their gating and ion permeation properties. Here we report a high-resolution cryo-electron microscopy structure of the mouse Piezo1 trimer. The detergent-solubilized complex adopts a three-bladed propeller shape with a curved transmembrane region containing at least 26 transmembrane helices per protomer. The flexible propeller blades can adopt distinct conformations, and consist of a series of four-transmembrane helical bundles that we term Piezo repeats. Carboxy-terminal domains line the central ion pore, and the channel is closed by constrictions in the cytosol. A kinked helical beam and anchor domain link the Piezo repeats to the pore, and are poised to control gating allosterically. The structure provides a foundation to dissect further how Piezo channels are regulated by mechanical force.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
04 July 2022
A Correction to this paper has been published: https://doi.org/10.1038/s41586-022-04976-8
References
Ranade, S. S., Syeda, R. & Patapoutian, A. Mechanically activated ion channels. Neuron 87, 1162–1179 (2015)
Coste, B. et al. Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science 330, 55–60 (2010)
Coste, B. et al. Piezo proteins are pore-forming subunits of mechanically activated channels. Nature 483, 176–181 (2012)
Ranade, S. S. et al. Piezo2 is the major transducer of mechanical forces for touch sensation in mice. Nature 516, 121–125 (2014)
Woo, S. H. et al. Piezo2 is the principal mechanotransduction channel for proprioception. Nat. Neurosci. 18, 1756–1762 (2015)
Kim, S. E., Coste, B., Chadha, A., Cook, B. & Patapoutian, A. The role of Drosophila Piezo in mechanical nociception. Nature 483, 209–212 (2012)
Li, J. et al. Piezo1 integration of vascular architecture with physiological force. Nature 515, 279–282 (2014)
Nonomura, K. et al. Piezo2 senses airway stretch and mediates lung inflation-induced apnoea. Nature 541, 176–181 (2017)
Zarychanski, R. et al. Mutations in the mechanotransduction protein PIEZO1 are associated with hereditary xerocytosis. Blood 120, 1908–1915 (2012)
Albuisson, J. et al. Dehydrated hereditary stomatocytosis linked to gain-of-function mutations in mechanically activated PIEZO1 ion channels. Nat. Commun. 4, 1884 (2013)
Andolfo, I. et al. Multiple clinical forms of dehydrated hereditary stomatocytosis arise from mutations in PIEZO1. Blood 121, 3925–3935 (2013)
Bae, C., Gnanasambandam, R., Nicolai, C., Sachs, F. & Gottlieb, P. A. Xerocytosis is caused by mutations that alter the kinetics of the mechanosensitive channel PIEZO1. Proc. Natl Acad. Sci. USA 110, E1162–E1168 (2013)
Coste, B. et al. Gain-of-function mutations in the mechanically activated ion channel PIEZO2 cause a subtype of Distal Arthrogryposis. Proc. Natl Acad. Sci. USA 110, 4667–4672 (2013)
Lukacs, V. et al. Impaired PIEZO1 function in patients with a novel autosomal recessive congenital lymphatic dysplasia. Nat. Commun. 6, 8329 (2015)
Chesler, A. T. et al. The role of PIEZO2 in human mechanosensation. N. Engl. J. Med. 375, 1355–1364 (2016)
Mahmud, A. A. et al. Loss of the proprioception and touch sensation channel PIEZO2 in siblings with a progressive form of contractures. Clin. Genet. 91, 470–475 (2017)
Haliloglu, G. et al. Recessive PIEZO2 stop mutation causes distal arthrogryposis with distal muscle weakness, scoliosis and proprioception defects. J. Hum. Genet. 62, 497–501 (2017)
Syeda, R. et al. Piezo1 channels are inherently mechanosensitive. Cell Reports 17, 1739–1746 (2016)
Lewis, A. H. & Grandl, J. Mechanical sensitivity of Piezo1 ion channels can be tuned by cellular membrane tension. eLife 4, e12088 (2015)
Cox, C. D. et al. Removal of the mechanoprotective influence of the cytoskeleton reveals PIEZO1 is gated by bilayer tension. Nat. Commun. 7, 10366 (2016)
Kamajaya, A., Kaiser, J. T., Lee, J., Reid, M. & Rees, D. C. The structure of a conserved Piezo channel domain reveals a topologically distinct β sandwich fold. Structure 22, 1520–1527 (2014)
Ge, J. et al. Architecture of the mammalian mechanosensitive Piezo1 channel. Nature 527, 64–69 (2015)
Brohawn, S. G., Campbell, E. B. & MacKinnon, R. Physical mechanism for gating and mechanosensitivity of the human TRAAK K+ channel. Nature 516, 126–130 (2014)
Pliotas, C. et al. The role of lipids in mechanosensation. Nat. Struct. Mol. Biol. 22, 991–998 (2015)
Phillips, R., Ursell, T., Wiggins, P. & Sens, P. Emerging roles for lipids in shaping membrane-protein function. Nature 459, 379–385 (2009)
Jasti, J., Furukawa, H., Gonzales, E. B. & Gouaux, E. Structure of acid-sensing ion channel 1 at 1.9 Å resolution and low pH. Nature 449, 316–323 (2007)
Kawate, T., Michel, J. C., Birdsong, W. T. & Gouaux, E. Crystal structure of the ATP-gated P2X4 ion channel in the closed state. Nature 460, 592–598 (2009)
Zhao, Q. et al. Ion permeation and mechanotransduction mechanisms of mechanosensitive Piezo channels. Neuron 89, 1248–1263 (2016)
Long, S. B., Campbell, E. B. & Mackinnon, R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science 309, 897–903 (2005)
Liao, M., Cao, E., Julius, D. & Cheng, Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature 504, 107–112 (2013)
Coste, B. et al. Piezo1 ion channel pore properties are dictated by C-terminal region. Nat. Commun. 6, 7223 (2015)
Drin, G. & Antonny, B. Amphipathic helices and membrane curvature. FEBS Lett. 584, 1840–1847 (2010)
Bavi, N. et al. The role of MscL amphipathic N terminus indicates a blueprint for bilayer-mediated gating of mechanosensitive channels. Nat. Commun. 7, 11984 (2016)
Mansoor, S. E. et al. X-ray structures define human P2X3 receptor gating cycle and antagonist action. Nature 538, 66–71 (2016)
Wu, J., Lewis, A. H. & Grandl, J. Touch, tension, and transduction - the function and regulation of Piezo ion channels. Trends Biochem. Sci. 42, 57–71 (2017)
Glogowska, E. et al. Novel mechanisms of PIEZO1 dysfunction in hereditary xerocytosis. Blood 130, 1845–1856 (2017)
Wu, J., Goyal, R. & Grandl, J. Localized force application reveals mechanically sensitive domains of Piezo1. Nat. Commun. 7, 12939 (2016)
Ursell, T., Kondev, J., Reeves, D., Wiggins, P. & Phillips, R. in Mechanosensitive Ion Channels Vol. 1 (eds Kamkin, A. & Kiseleva, I. ) 37–70 (Springer, 2008)
Suloway, C. et al. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151, 41–60 (2005)
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017)
Zhang, K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016)
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012)
Berndsen, Z., Bowman, C., Jang, H. & Ward, A. B. EMHP: An accurate automated hole masking algorithm for single-particle cryo-EM image processing. Bioinformatics 33, 3823–3826 (2017)
Bartesaghi, A., Matthies, D., Banerjee, S., Merk, A. & Subramaniam, S. Structure of beta-galactosidase at 3.2-Å resolution obtained by cryo-electron microscopy. Proc. Natl Acad. Sci. USA 111, 11709–11714 (2014)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
Adams, P. D . et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010)
Barad, B. A. et al. EMRinger: side chain directed model and map validation for 3D cryo-electron microscopy. Nat. Methods 12, 943–946 (2015)
Unni, S. et al. Web servers and services for electrostatics calculations with APBS and PDB2PQR. J. Comput. Chem. 32, 1488–1491 (2011)
The PyMOL Molecular Graphics System. Version 1.8 Schrödinger, LLC; http://www.pymol.org/ (2015)
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004)
Acknowledgements
We thank H. Turner and G. Ozorowski for assistance with electron microscopy data collection. We acknowledge early efforts to characterize Piezo1 using electron microscopy by E. Wilson-Kubalek and R. Milligan, and S. Kakuda for screening purification conditions. We thank A. Sobolevsky for critical reading of the manuscript, and members of the Ward and Patapoutian laboratories for discussion. This work was supported by a Ray Thomas Edwards Foundation grant to A.B.W. and National Institutes of Health (NIH) grants NS083174 and DE022358 to A.P. Computational analyses were performed using shared instrumentation funded by NIH 1-S10OD021634. A.P. is an investigator of the Howard Hughes Medical Institute. This is manuscript 29582 from The Scripps Research Institute.
Author information
Authors and Affiliations
Contributions
K.S. prepared electron microscopy samples, collected and processed electron microscopy data, built the structural model and conducted fluorescence-detection size-exclusion chromatography analysis. S.E.M. carried out electrophysiological experiments and data analysis. J.M.K. developed sample preparation protocols. T.W. carried out immunostaining experiments. A.P. and A.B.W. supervised the project. K.S. drafted the manuscript, which was edited by A.P. and A.B.W. with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 Purification and electron microscopy analysis of Piezo1.
a, Preparative gel filtration chromatogram of mouse Piezo1 after affinity purification and proteolytic removal of the GST tag. b, SDS–PAGE analysis of Piezo1 following affinity purification (pre-SEC) and after subsequent size-exclusion chromatography (SEC) steps (SEC fractions). Purifications of Piezo1 have been repeated more than three times with similar results. c, Aligned micrograph of purified Piezo1 embedded in a thin layer of vitrified ice. Scale bar, 100?nm. d, Representative 2D classes of Piezo1 showing different particle orientations.
Extended Data Figure 2 Classification and refinement of Piezo1 core.
a, Data processing flow chart. b, Local resolution maps calculated by the locres program in RELION 2.0. Colour key for local resolution (in Å) is shown. c, Angular distributions of the particles after the final step of refinement in RELION. The radius of the sphere is proportional to the number of particles with a given orientation. Only one-third of the sphere is shown due to applied C3 symmetry. d, FSC plots calculated using relion_postprocess for unmasked maps and maps with a soft mask applied to remove the contribution of scattered detergent density.
Extended Data Figure 3 Asymmetry of Piezo1 and blade-focused classification and refinement.
a, Duplicate unsharpened maps of Piezo1 refined without symmetry imposed, with the blue map rotated approximately 120° around the central axis of pseudosymmetry relative to the pink map, then superimposed with the ‘fit in map’ function of UCSF Chimera. The left panel shows top view, while the right panel shows a horizontal slice through the transmembrane region. b, Flow chart of propeller blade-focused classification and refinement procedure. c, Local resolution maps calculated by the locres program in RELION 2.0. Colour key for local resolution (in Å) is shown and is the same as Extended Data Fig. 2b. d, Angular distributions of the particles after the final step of refinement in RELION. The radius of the sphere is proportional to the number of particles with a given orientation. e, FSC plots calculated using relion_postprocess for unmasked maps and maps with a soft mask applied to remove the contribution of scattered detergent density.
Extended Data Figure 4 Fit of molecular model to electron density.
Select regions of the molecular model are shown as yellow cartoon, with superimposed electron density as blue mesh. The density is derived from the C3-symmetry core masked map for the top two rows, and derived from blade class 1 for the bottom row.
Extended Data Figure 5 Structural comparisons of current and previous (EMDB 6343) mouse Piezo1 structures.
For each panel, the ‘fit in map’ function of UCSF Chimera was used to align uncropped maps. In a–e, unsharpened map of C3 refinement is shown. a–c, Top (a), bottom (b) and side (c) views of current Piezo1 (green) and EMDB accession EMD-6343 (grey) structurally aligned. d, Expanded top view of the cap domain to illustrate an approximately 15° rotation of the cap between the two maps. e, Expanded sliced top view of pore-proximal transmembrane region. Subtle conformational rearrangements are present between the current and previous structures in the inner and outer helices. f, g, Top and side views of EMDB accession EMD-6343 (grey), blade class 1 (blue), and blade class 2 (orange). The trajectories of the propeller blades are broadly similar across the maps in both the membrane-parallel and membrane-normal directions.
Extended Data Figure 6 Structural features of Piezo repeats.
a, Top view of Piezo1 depicted as cartoon, with amphipathic helices from Piezo repeats A, B and C shown in red. Extracellular cap domain is omitted for clarity. b–d, Ribbon models of amphipathic helices from Piezo repeat A (b), Piezo repeat B (c), and Piezo repeat C (d). Cα positions are shown as spheres and hydrophobic amino acids are coloured grey, basic amino acids are coloured blue, acidic amino acids are coloured red, and polar uncharged amino acids are coloured yellow. e, Cartoon model of a portion of the Piezo1 propeller blade, with Piezo repeats coloured separately. A strong density peak putatively representing a lipid head group is shown as a pink mesh. f, Expanded view of the putative lipid binding site sandwiched between Piezo repeats B and C (helices B4 and C1). Electron density (blue mesh) is superimposed onto the molecular model, and the putative lipid head group is shown as pink mesh. Side chains contributing to the binding pocket are shown as yellow sticks and labelled.
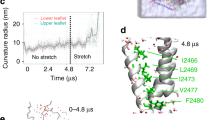
Extended Data Figure 7 Expression and functional properties of Piezo1 mutants.
a, HEK293T cells deficient in Piezo1 (P1KO) that express the Piezo1 double mutant M2493A/F2494A do not show discernible currents activated by indentation with blunt pipette (left) or stretch (right). b, Fluorescence-detection size-exclusion chromatography traces of untransfected HEK293F cells (grey) or cells expressing C-terminal tdTomato fusions of wild-type Piezo1 or double mutant M2493A/F2494A injected into Superose 6 increase column. Similar to the wild type, the double mutant displays a single dominant peak eluting shortly after void volume, indicating proper trimeric expression. Data are from one independent experiment. c, Representative images of Myc labelling in HEK293T-P1KO cells transfected with Piezo1–IRES-GFP (top), Piezo1–Myc–IRES-GFP (middle) or Piezo1(M2493A/F2494A)–Myc–IRES-GFP (bottom). GFP, green fluorescent protein. IRES, internal ribozyme entry site. Myc tags were inserted at position 897. Immunostaining was performed before (left) or after (right) cell permeabilization. Piezo1(M2493A/F2494A)–Myc is labelled at the surface in live cells similar to Piezo1–Myc, indicating that surface expression is preserved in the non-functional mutant. Scale bar, 10?μm. Experiments were repeated three times with reproducible results. d, Left, representative traces of stretch-activated single channel currents recorded (−80?mV) from HEK293T-P1KO cells expressing wild-type, M2493A or F2494A Piezo1 channels (traces displayed after applying 1?kHz digital filter). The corresponding pressure stimulus used to elicit the response is illustrated above the current trace. For each condition, the amplitude histogram for the corresponding trace is depicted on the right. The Gaussian fit for the closed (black curve) and open (red curve) components is overlaid on the histograms. e, Left, average I–V relationship of stretch-activated single-channel currents from wild-type (n?=?4), M2493A (n?=?5) or F2494A (n?=?4) Piezo1 channels. Amplitude was measured as a difference in Gaussian fits of full-trace histograms. Right, mean unitary conductance calculated from the slope of linear regression line fit to individual cells in each condition. **P?<?0.01, one-way ANOVA with Dunn’s comparison. f, Mean I–V relationship curves of mechanically activated currents recorded from wild-type (n?=?6), M2493A (n?=?8) or F2494A (n?=?9) Piezo1 channels with 150?mM CsCl-based intracellular solution and 100?mM CaCl2-based extracellular solution. Currents were elicited from −69.6 to 50.4?mV (Δ20?mV). Inset, expanded view of curves between 5 and 15?mV. Values are mean?±?s.e.m. g, Mean reversal potential from individual cells; Piezo1: 8.8?±?1.3?mV (n?=?6), M2493A: 10.2?±?1.2?mV (n?=?8) and F2494A: 13.5?±?0.5?mV (n?=?9). **P?=?0.0063, one-way ANOVA with Dunn’s multiple comparison test. N denotes X individual cells.
Supplementary information
Supplementary Data
Sequence alignment of Piezo orthologues. Amino acid sequence alignment of selected regions of mouse Piezo1 (mPiezo1; UniProt E2JF22), human Piezo1 (hPiezo1; UniProt Q92508), mouse Piezo2 (mPiezo2, UniProt Q8CD54), human Piezo2 (hPiezo2, UniProt Q9H5I5), fruit fly Piezo (dmPiezo, UniProt M9MSG8). Structural features are annotated above the mPiezo1 sequence. Unmodeled regions are depicted as dotted lines. (PDF 4081 kb)
Rights and permissions
About this article
Cite this article
Saotome, K., Murthy, S., Kefauver, J. et al. Structure of the mechanically activated ion channel Piezo1. Nature 554, 481–486 (2018). https://doi.org/10.1038/nature25453
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature25453
This article is cited by
-
Bioelectricity in dental medicine: a narrative review
BioMedical Engineering OnLine (2024)
-
PIEZO1 loss-of-function compound heterozygous mutations in the rare congenital human disorder Prune Belly Syndrome
Nature Communications (2024)
-
Piezo1, the new actor in cell volume regulation
Pflügers Archiv - European Journal of Physiology (2024)
-
Piezo2 Contributes to Traumatic Brain Injury by Activating the RhoA/ROCK1 Pathways
Molecular Neurobiology (2024)
-
The functional effects of Piezo channels in mesenchymal stem cells
Stem Cell Research & Therapy (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.