Abstract
Here we describe the honeycomb maze, a behavioural paradigm for the study of spatial navigation in rats. The maze consists of 37 platforms that can be raised or lowered independently. Place navigation requires an animal to go to a goal platform from any of several start platforms via a series of sequential choices. For each, the animal is confined to a raised platform and allowed to choose between two of the six adjacent platforms, the correct one being the platform with the smallest angle to the goal-heading direction. Rats learn rapidly and their choices are influenced by three factors: the angle between the two choice platforms, the distance from the goal, and the angle between the correct platform and the direction of the goal. Rats with hippocampal damage are impaired in learning and their performance is affected by all three factors. The honeycomb maze represents a marked improvement over current spatial navigation tests, such as the Morris water maze1,2,3, because it controls the choices of the animal at each point in the maze, provides the ability to assess knowledge of the goal direction from any location, enables the identification of factors influencing task performance and provides the possibility for concomitant single-cell recording.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Morris, R. G. M. Spatial localization does not require the presence of local cues. Learn. Motiv. 12, 239–260 (1981)
Morris, R. G., Garrud, P., Rawlins, J. N. & O’Keefe, J. Place navigation impaired in rats with hippocampal lesions. Nature 297, 681–683 (1982)
Morris, R. Developments of a water-maze procedure for studying spatial learning in the rat. J. Neurosci. Methods 11, 47–60 (1984)
O’Keefe, J. & Nadel, L. The Hippocampus as a Cognitive Map (Oxford Univ. Press, 1978)
O’Keefe, J. & Dostrovsky, J. The hippocampus as a spatial map. Preliminary evidence from unit activity in the freely-moving rat. Brain Res. 34, 171–175 (1971)
Ranck, J. B. Head-direction cells in the deep cell layers of the dorsal presubiculum in freely moving rats. Abstr. Soc. Neurosci. 10, 599 (1984)
Taube, J. S., Muller, R. U. & Ranck, J. B. Jr. Head-direction cells recorded from the postsubiculum in freely moving rats. I. Description and quantitative analysis. J. Neurosci. 10, 420–435 (1990)
Hafting, T., Fyhn, M., Molden, S., Moser, M. B. & Moser, E. I. Microstructure of a spatial map in the entorhinal cortex. Nature 436, 801–806 (2005)
Lever, C., Burton, S., Jeewajee, A., O’Keefe, J. & Burgess, N. Boundary vector cells in the subiculum of the hippocampal formation. J. Neurosci. 29, 9771–9777 (2009)
Solstad, T., Boccara, C. N., Kropff, E., Moser, M. B. & Moser, E. I. Representation of geometric borders in the entorhinal cortex. Science 322, 1865–1868 (2008)
O’Keefe, J. in The Hippocampus Book (eds Morris, R. et al.) Ch. 8, 471–544 (Oxford Univ. Press, 2007)
Moser, M. B., Rowland, D. C. & Moser, E. I. Place cells, grid cells, and memory. Cold Spring Harb. Perspect. Biol. 7, a021808 (2015)
Dudchenko, P. A. How do animals actually solve the T maze? Behav. Neurosci. 115, 850–860 (2001)
Olton, D. S., Walker, J. A. & Gage, F. H. Hippocampal connections and spatial discrimination. Brain Res. 139, 295–308 (1978)
Olton, D. S. & Samuelson, R. J. Remembrance of places passed: spatial memory in rats. J. Exp. Psychol. Anim. Behav. Process. 2, 97–116 (1976)
Barnes, C. A. Memory deficits associated with senescence: a neurophysiological and behavioral study in the rat. J. Comp. Physiol. Psychol. 93, 74–104 (1979)
O’Keefe, J. in Brain and Space (ed. Paillard, J. ) 273–295 (Oxford Univ. Press, 1991)
O’Keefe, J. in Language and Space (eds Bloom, P. et al.) 277–316 (MIT Press, 1996)
Eichenbaum, H., Stewart, C. & Morris, R. G. Hippocampal representation in place learning. J. Neurosci. 10, 3531–3542 (1990)
Acknowledgements
We thank M. Bertelli, A. Hastings, D. Howett, N. Khan, P. Mumford, A. O’Leary, B. Potter, S. Richards and R. Wu for their contribution to this work, and D. Farquharson and D. Halpin for their input to the design, building and testing of maze prototypes. This work was supported by grants from the Wellcome Trust and the Gatsby Charitable Foundation to J.O. R.A.W. is an MRC Clinical Research Training Fellow, J.K. is a Wellcome Trust/Royal Society Sir Henry Dale Fellow and is supported by the Kavli Foundation Dream Team project and the Isaac Newton Trust. D.C. is funded by the Cambridge NIHR Biomedical Research Centre and by the Wellcome Trust.
Author information
Authors and Affiliations
Contributions
J.O. conceived the maze and the study. J.O., M.B., J.K. and S.B. were instrumental in designing, building and testing prototypes of the maze. J.K. and M.B. designed the custom-made software used to operate the maze. R.A.W. designed testing schedule 1 for the control experiment, and A.D. and J.O. designed testing schedule 2 for the lesion experiment. A.D. and S.B. performed the hippocampal lesion and sham lesion surgeries. R.A.W. acquired the behavioural data. R.A.W. and A.D. performed the histology for the lesion experiment, and R.A.W. measured hippocampal lesion volumes. R.A.W. conducted the data management and performed the statistical analyses. J.K., S.B. and J.O. collected the single-unit data and J.K. and J.O. analysed these data. J.O. and R.A.W. wrote the manuscript, with contributions to later drafts from all other authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
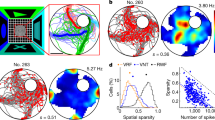
Extended Data Figure 1 Place cell recorded on the honeycomb maze.
A single place cell recorded during navigation on the honeycomb maze. a, Behaviour (black line) from a single trial (trial i) in which the rat was offered two consecutive choices (left, 0–73 s) to go from the start platform (second left, 0–25 s), to an intermediate platform (third left, 25–50 s) to the goal (right, 50–73 s). The program detained the rat on each platform for 20 s before the two choice platforms were raised or, in goal, the food presented. Non-chosen platforms not shown. As the rat waited on each platform, it sampled the immediate environment by circling the perimeter of the platform. b, The firing of a place cell during this trial (trial i); maximum rate in red shown top left. c, Rate map for the same cell on a separate trial (trial g) from a different start platform. d, Composite rate map from ten trials (trials c–l), each from a different starting location in which the rat took a different path to the goal. e, Firing rate map of the same cell when all platforms were raised and the rat foraged for food over a period of 20 min (trial m). In this cell, the firing fields during navigation trials (d) and during the foraging condition (e) were similar (spatial correlation = 0.77). Not all place cells displayed this profile, and others (not shown) fired in a different location(s) during the navigation trials from that seen in the foraging condition (that is, remapped).
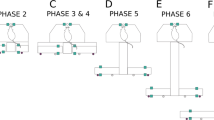
Extended Data Figure 2 Protocols used on the honeycomb maze.
a, Schedule 1 trial protocols for Aβ60 (left), Aβ120 (middle) and Aβ180 (right) trials. b, Schedule 2 protocol. For a and b: goal platform, black; start platforms, blue; orange vectors, correct choices; grey vectors, incorrect choices; green vectors, ‘forced’ choices. See Methods for more detail.
Extended Data Figure 3 Histology of brains from rats with hippocampal lesions.
Representative sections from brains of rats with hippocampal lesions, and one operated control with a sham hippocampal lesion (R2322), alongside mean performance scores on the honeycomb maze. Subjects are arranged in order of increasing lesion size. Horizontal sections (40 μm) stained with cresyl violet.
Extended Data Figure 4 Correlation between hippocampal volume and performance.
Correlation between remaining hippocampal volume and performance on the spatial navigation task on the honeycomb maze in eight rats with hippocampal lesions (n = 8 rats, ρ6 = 0.452, P = 0.260; Spearman’s correlation).
Extended Data Figure 5 Rats with hippocampal lesions have longer latencies.
Rats with hippocampal lesions (red, n = 8) have longer latencies than operated controls with sham hippocampal lesions (blue, n = 8) (F1,14 = 11.103, P = 0.005). Latencies also changed as a function of experience (day) (F16,224 = 5.612, P < 0.001) with a significant day × lesion interaction (F16,224 = 2.464, P = 0.002, two-way mixed ANOVA). **P < 0.005. Error bars indicate s.e.m.
Extended Data Figure 6 Vector-based navigation schema.
Left, The hippocampus represents a goal-direction vector pointing from the rat to the goal (A), which decreases as the rat is farther from the goal (right). The navigation system computes the projection of each choice platform vector (B, C) onto the goal-direction vector (inner product, Bgd, Cgd) and selects the larger of the two. This choice is easier with increased angle between choices (angle β) and consequent increased difference in the magnitudes of their projection vectors. The projection vector of the preferred platform, Bgd, is the output of the system that competes with other potential solutions to the problem (for example, choose between the leftmost or northmost platform).
Supplementary information
Supplementary Information
This file contains a Supplementary Discussion and additional references. (PDF 262 kb)
Control rat navigating the Honeycomb Maze
A control rat making a series of choices as it navigates to the goal on the Honeycomb Maze. (MP4 5684 kb)
Rights and permissions
About this article
Cite this article
Wood, R., Bauza, M., Krupic, J. et al. The honeycomb maze provides a novel test to study hippocampal-dependent spatial navigation. Nature 554, 102–105 (2018). https://doi.org/10.1038/nature25433
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature25433
This article is cited by
-
Rodent maze studies: from following simple rules to complex map learning
Brain Structure and Function (2024)
-
Hippocampal place cells have goal-oriented vector fields during navigation
Nature (2022)
-
The art gallery maze: a novel tool to assess human navigational abilities
Cognitive Processing (2021)
-
High-Throughput Automatic Training System for Spatial Working Memory in Free-Moving Mice
Neuroscience Bulletin (2019)
-
A micro-CT-based method for quantitative brain lesion characterization and electrode localization
Scientific Reports (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.