Abstract
Inflammatory diseases of the gastrointestinal tract are frequently associated with dysbiosis1,2,3,4,5,6,7,8, characterized by changes in gut microbial communities that include an expansion of facultative anaerobic bacteria of the Enterobacteriaceae family (phylum Proteobacteria). Here we show that a dysbiotic expansion of Enterobacteriaceae during gut inflammation could be prevented by tungstate treatment, which selectively inhibited molybdenum-cofactor-dependent microbial respiratory pathways that are operational only during episodes of inflammation. By contrast, we found that tungstate treatment caused minimal changes in the microbiota composition under homeostatic conditions. Notably, tungstate-mediated microbiota editing reduced the severity of intestinal inflammation in mouse models of colitis. We conclude that precision editing of the microbiota composition by tungstate treatment ameliorates the adverse effects of dysbiosis in the inflamed gut.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
References
Frank, D. N. et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl Acad. Sci. USA 104, 13780–13785 (2007)
Lupp, C. et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2, 119–129 (2007)
Garrett, W. S. et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe 8, 292–300 (2010)
Mshvildadze, M. et al. Intestinal microbial ecology in premature infants assessed with non-culture-based techniques. J. Pediatr. 156, 20–25 (2010)
Vujkovic-Cvijin, I. et al. Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci. Transl. Med. 5, 193ra91 (2013)
Raetz, M. et al. Parasite-induced TH1 cells and intestinal dysbiosis cooperate in IFN-γ-dependent elimination of Paneth cells. Nat. Immunol. 14, 136–142 (2013)
Winter, S. E., Lopez, C. A. & Bäumler, A. J. The dynamics of gut-associated microbial communities during inflammation. EMBO Rep. 14, 319–327 (2013)
Shin, N. R., Whon, T. W. & Bae, J. W. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 33, 496–503 (2015)
Garrett, W. S. et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell 131, 33–45 (2007)
Elinav, E. et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell 145, 745–757 (2011)
Hughes, E. R. et al. Microbial respiration and formate oxidation as metabolic signatures of inflammation-associated dysbiosis. Cell Host Microbe 21, 208–219 (2017)
Winter, S. E. et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 339, 708–711 (2013)
Gates, A. J. et al. Properties of the periplasmic nitrate reductases from Paracoccus pantotrophus and Escherichia coli after growth in tungsten-supplemented media. FEMS Microbiol. Lett. 220, 261–269 (2003)
Hans, W., Schölmerich, J., Gross, V. & Falk, W. The role of the resident intestinal flora in acute and chronic dextran sulfate sodium-induced colitis in mice. Eur. J. Gastroenterol. Hepatol. 12, 267–273 (2000)
Rutgeerts, P. et al. Controlled trial of metronidazole treatment for prevention of Crohn’s recurrence after ileal resection. Gastroenterology 108, 1617–1621 (1995)
Prantera, C. et al. An antibiotic regimen for the treatment of active Crohn’s disease: a randomized, controlled clinical trial of metronidazole plus ciprofloxacin. Am. J. Gastroenterol. 91, 328–332 (1996)
Stecher, B. et al. Salmonella enterica serovar Typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 5, 2177–2189 (2007)
Collins, S., Verdu, E., Denou, E. & Bercik, P. The role of pathogenic microbes and commensal bacteria in irritable bowel syndrome. Dig. Dis. 27 (Suppl 1), 85–89 (2009)
Kamada, N. et al. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science 336, 1325–1329 (2012)
Brugiroux, S. et al. Genome-guided design of a defined mouse microbiota that confers colonization resistance against Salmonella enterica serovar Typhimurium. Nat. Microbiol. 2, 16215 (2016)
Kruis, W. et al. Maintaining remission of ulcerative colitis with the probiotic Escherichia coli Nissle 1917 is as effective as with standard mesalazine. Gut 53, 1617–1623 (2004)
Sassone-Corsi, M. et al. Microcins mediate competition among Enterobacteriaceae in the inflamed gut. Nature 540, 280–283 (2016)
Sambrook, J ., Fritsch, E. F. & Maniatis, T. Molecular Cloning 2nd edn (Cold Spring Harbor Laboratory Press, 1989)
Davis, R. W ., Roth, J. R. & Botstein, D. Advanced Bacterial Genetics (Cold Spring Harbor Laboratory Press, 1980)
Price-Carter, M., Tingey, J., Bobik, T. A. & Roth, J. R. The alternative electron acceptor tetrathionate supports B12-dependent anaerobic growth of Salmonella enterica serovar Typhimurium on ethanolamine or 1,2-propanediol. J. Bacteriol. 183, 2463–2475 (2001)
Caporaso, J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010)
Shavit, Y., Hamey, F. K. & Lio, P. FisHiCal: an R package for iterative FISH-based calibration of Hi-C data. Bioinformatics 30, 3120–3122 (2014)
Li, W. & Godzik, A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659 (2006)
Fu, L., Niu, B., Zhu, Z., Wu, S. & Li, W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152 (2012)
Barman, M. et al. Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infect. Immun. 76, 907–915 (2008)
Winter, S. E. et al. Contribution of flagellin pattern recognition to intestinal inflammation during Salmonella enterica serotype Typhimurium infection. Infect. Immun. 77, 1904–1916 (2009)
Winter, S. E. et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467, 426–429 (2010)
Bacchetti De Gregoris, T., Aldred, N., Clare, A. S. & Burgess, J. G. Improvement of phylum- and class-specific primers for real-time PCR quantification of bacterial taxa. J. Microbiol. Methods 86, 351–356 (2011)
Stewart, V. & Parales, J. Jr. Identification and expression of genes narL and narX of the nar (nitrate reductase) locus in Escherichia coli K-12. J. Bacteriol. 170, 1589–1597 (1988)
Rodriguez, M. S., Thompson, J., Hay, R. T. & Dargemont, C. Nuclear retention of IκBα protects it from signal-induced degradation and inhibits nuclear factor κB transcriptional activation. J. Biol. Chem. 274, 9108–9115 (1999)
Keestra, A. M. et al. A Salmonella virulence factor activates the NOD1/NOD2 signaling pathway. MBio 20, e00266–11 (2011)
Eaves-Pyles, T. et al. Escherichia coli isolated from a Crohn’s disease patient adheres, invades, and induces inflammatory responses in polarized intestinal epithelial cells. Int. J. Med. Microbiol. 298, 397–409 (2008)
Kim, S. C. et al. Variable phenotypes of enterocolitis in interleukin 10-deficient mice monoassociated with two different commensal bacteria. Gastroenterology 128, 891–906 (2005)
Pal, D., Venkova-Canova, T., Srivastava, P. & Chattoraj, D. K. Multipartite regulation of rctB, the replication initiator gene of Vibrio cholerae chromosome II. J. Bacteriol. 187, 7167–7175 (2005)
Simon, R., Priefer, U. & Puhler, A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat. Biotechnol. 1, 784–791 (1983)
Overbergh, L. et al. The use of real-time reverse transcriptase PCR for the quantification of cytokine gene expression. J. Biomol. Tech. 14, 33–43 (2003)
Godinez, I. et al. T cells help to amplify inflammatory responses induced by Salmonella enterica serotype Typhimurium in the intestinal mucosa. Infect. Immun. 76, 2008–2017 (2008)
Wilson, R. P. et al. The Vi-capsule prevents Toll-like receptor 4 recognition of Salmonella. Cell. Microbiol. 10, 876–890 (2008)
Wang, R. F. & Kushner, S. R. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100, 195–199 (1991)
Acknowledgements
This work was supported by NIH grants AI112445 (A.J.B.), AI118807 (S.E.W.), AI128151 (S.E.W.), DK070855 (L.V.H.), DK102436 (B.A.D.) and 5K12HD-068369 (L.S.-D.), Welch Foundation grants I-1858 (S.E.W.) and I-1874 (L.V.H.), American Cancer Society Research Scholar Grant MPC-130347 (S.E.W.) and a Crohn’s and Colitis Foundation of America postdoctoral fellowship no. 454921 (W.Z.). Work in the L.V.H. laboratory is supported by the Howard Hughes Medical Institute. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the funders. We thank B. Sartor for the E. coli NC101 strain.
Author information
Authors and Affiliations
Contributions
W.Z., M.G.W., L.S., L.B., E.d.L.R., R.P.W., E.R.H, C.A.L. and C.C.G. performed and analysed nitrate reductase activity, in vitro bacterial competitive growth, NF-κB induction, DSS cytotoxity experiments and experiments involving conventionally raised C57BL/6 mice. M.G.W., L.S. and E.R.H. performed Il10−/− mouse experiments. W.Z. and M.G.W. performed inflammation analysis. C.L.B., M.G.W., L.S. and W.Z. performed germ-free-mouse experiments. B.A.D. and W.Z. analysed 16S and metagenomic data. M.X.B. analysed the histopathology. L.S.-D. and K.H.-H. contributed to humanized mouse experiments. L.S. and S.E.W. performed metabolite quantification. W.Z., Ç.T., A.Y.K., E.B., L.V.H., A.J.B. and S.E.W. designed the experiments, interpreted the data and wrote the manuscript with contributions from all authors.
Corresponding authors
Ethics declarations
Competing interests
A patent application has been filed on behalf of A.J.B. and S.E.W. and The Regents Of The University Of California based on some of the results reported in this letter (Application number, US 14/964,487). All other authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks C. Elson, M. Fischbach and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
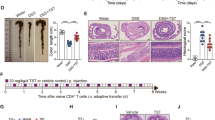
Extended Data Figure 1 Effect of tungstate on anaerobic respiration in vitro.
a, Nitrate reductase activity of E. coli Nissle 1917 measured in medium supplemented with sodium nitrate and the indicated concentrations of sodium tungstate. b, Competitive growth of the E. coli Nissle 1917 wild-type strain and the isogenic molybdenum-cofactor-deficient mutant in the presence of the indicated electron acceptors under anaerobic conditions. c, Competitive growth of E. coli NRG857c wild-type strain and the isogenic molybdenum-cofactor-deficient mutant in the presence of the indicated electron acceptors under anaerobic conditions. In a–c, n = 3 replicates per condition; n denotes the number of biological replicates. Data are shown as geometric mean and geometric s.d. of three experiments.
Extended Data Figure 2 Tungstate treatment of mice colonized with E. coli K-12.
Conventionally raised C57BL/6 mice were treated with 0.2% sodium tungstate, DSS, DSS plus sodium tungstate or left untreated (mock). After four days, animals were inoculated orally with the E. coli K-12 wild-type strain and the isogenic moaA mutant. C57BL/6 mice with naive microbiota (endogenous Enterobacteriaceae only) or germ-free C57BL/6 mice were treated similarly but were not inoculated with E. coli indicator strains. Samples were analysed after a total of nine days of treatment. a, b, Schematic representations of the colitis models. c–h, Transcription of Nos2 (c), Tnf (d), Il6 (e), Lcn2 (f), Cxcl2 (g) and Ifng (h) in the caecal mucosa was determined by RT–qPCR. E. coli K-12: n = 6 per group. Endogenous Enterobacteriaceae: mock, n = 14; W, n = 6; DSS, n = 19; DSS+W, n = 19. Germ-free: mock, n = 5; DSS, n = 4; DSS+W, n = 5. i, Cumulative histopathology score for the colon tissue; mock, n = 14; W, n = 6; DSS, n = 19; DSS+W, n = 19; data are shown as mean and s.e.m. and each dot represents one animal. j, Colon length, n = 8 per group. k, Body weight of mice harbouring endogenous Enterobacteriaceae, n = 8 per group. l, Body weight of mice experimentally colonized with E. coli K-12, n = 6 per group. Unless otherwise noted, data are shown as geometric mean and geometric s.d.
Extended Data Figure 3 Effect of tungstate treatment on mice experimentally colonized with E. coli Nissle 1917.
Groups of conventionally raised C57BL/6 mice were orally inoculated with the E. coli Nissle 1917 wild-type strain and treated with 0.2% sodium tungstate, DSS, DSS and sodium tungstate, or left untreated (mock) for nine days. a, Schematic representation of colitis model used in this figure. b, Bacterial load in the caecum (white bars) and colon content (grey bars). c, Formalin-fixed, haematoxylin and eosin-stained sections of the caecum were scored for the presence of inflammatory lesions; representative images of stained caecal sections. d, Cumulative histopathology score for the caecum tissue; data are shown as mean and s.e.m., and each dot represents one animal. In b–d, mock and tungsten, n = 11 per group; DSS and DSS+W, n = 15 per group. e, Animal body weight, n = 8 per group. f–h, Transcription of the inflammatory marker genes Cxcl1 (f), Nos2, (g) and Tnf (h) in the caecal mucosa was determined by RT–qPCR, n = 11 per group. Unless otherwise noted, data are shown as geometric mean and geometric s.d.
Extended Data Figure 4 Effect of tungstate treatment on mice experimentally colonized with Enterobacter cloacae and adherent invasive E. coli.
a–h, Conventionally raised C57BL/6 mice were treated with DSS or DSS plus tungstate. After four days, animals were inoculated intragastrically with the indicated bacterial strains. Samples were collected five days after inoculation. a, Schematic representation of the experiments. b, c, The total population of E. cloacae (b) and NRG857c (c; DSS, n = 12; DSS+W, n = 10) in the large intestinal content was determined by plating on selective medium. d, e, Animal body weight. In b and d: DSS, n = 8; DSS+W, n = 10. In e, n = 5 per group. f–h, Transcription levels of the inflammatory marker genes Cxcl1 (f), Nos2 (g) and Tnf (h) in the caecal mucosa were determined by RT–qPCR; DSS, n = 12; DSS+W, n = 10. i–m, Paired germ-free Swiss–Webster mice received human faecal transplants and were treated with DSS or DSS plus 0.2% sodium tungstate for seven days; DSS, n = 4; DSS+W, n = 4. i, Schematic representation of the experiments. j, The abundance of Enterobacteriaceae in the caecal content was determined by plating on selective medium (MacConkey agar). k, Formalin-fixed, haematoxylin and eosin-stained sections of the mouse caecum were scored for the presence of inflammatory lesions; l, representative images. m, Transcription levels of the inflammatory marker genes Nos2 and Il17 in the mouse caecal mucosa. Data are shown as geometric mean and geometric s.d.
Extended Data Figure 5 Effect of tungstate on the naive gut microbiome.
Groups of C57BL/6 mice naturally harbouring Enterobacteriaceae were treated with 0.2% sodium tungstate, DSS, DSS plus tungstate or mock treatment for nine days (see also Extended Data Fig. 3b). a, Relative abundance of genes involved in formate and nitrate utilization in the caecal content shown by shotgun metagenomic sequencing (MEGAN5). Each section of the pie chart is representative of the number of mapped reads obtained for the individual animals (n = 6 per group). b, Box-and-whisker plot (boxes show median, first and third quantiles, whisker denotes minimum to maximum range) of Chao1 alpha diversity of the caecal microbiota community based on 16S profiling (n = 6 per group). c, Abundance of endogenous Enterobacteriaceae family members determined by plating on selective medium (MacConkey agar, c): mock, n = 14; W, n = 6; DSS, n = 19; DSS+W, n = 19. Data are shown as geometric mean and geometric s.d.
Extended Data Figure 6 Effect of tungstate treatment on obligate anaerobic commensal bacteria.
a–c, Metagenomic analysis of the caecal content of mice described in Extended Data Fig. 3b. Principal coordinates analysis of global metabolic pathway (a) and quantification of reads involved in fumarate respiration (b) and butyrate production (c). Ellipses in a denote 95% confidence interval. Data are shown as mean and s.d; n = 6 per group. d–f, Bacteroides thetaiotaomicron or Bacteroides fragilis were cultured anaerobically in mucin broth at 37 °C for 48 h. The medium was supplemented with sodium tungstate or metronidazole as indicated. Succinate production by B. thetaiotaomicron (d) and B. fragilis (f) was assessed by GC–MS. The growth of B. thetaiotaomicron was quantified by plating serial dilutions of bacterial culture on blood agar (e). g, C. symbiosum was inoculated into chopped meat broth and incubated anaerobically at 37 °C for 36 h. Butyrate concentration in the medium was measured using GC–MS. h, C. symbiosum was cultured anaerobically in chopped meat broth at 37 °C for 48 h. The growth of C. symbiosum was determined by plating serial dilutions of bacterial culture on thioglycolate plates. n = 3 biological replicates per condition. Data are shown as geometric mean and geometric s.d. of three experiments.
Extended Data Figure 7 Assessment of overall health of mice treated orally with tungstate.
Groups of seven mice were either mock-treated or treated with 0.2% sodium tungstate in drinking water for nine days. a, Complete blood count. b, Serum alanine amino-transferase concentration. n = 7 animals per group. Data are shown as geometric mean and geometric s.d.
Extended Data Figure 8 Exposure of cultured cells to sodium tungstate.
a, Daily water consumption of mice inoculated with E. coli K-12. Each dot represents the average daily water consumption (ml per day) of three mice, obtained at eight time points, with two cages per treatment group, n = 16. b, HeLa57A cells, expressing luciferase under the control of a NF-κB-dependent promoter, were treated with PMA and sodium tungstate at the indicated concentrations. Relative luciferase activity was determined after 5 h. c, d, MODE-K or BMDMs cells were treated with DSS or DSS plus sodium tungstate at the indicated concentrations for 24 h. The release of lactate dehydrogenases into the culture supernatant by MODE-K cells (c) or BMDMs (d) was measured. In b–d, n = 3 biological replicates per condition. e, f, Groups of conventionally raised C57BL/6 mice were treated with DSS for four days. Animals were inoculated intragastrically with an equal mixture of the indicated E. coli Nissle 1917 wild-type strain and an isogenic moaA mutant. On the day of inoculation, a subset of mice was switched to DSS plus sodium tungstate for four days while a control group remained on DSS treatment. Schematic representation of experiment (e). The competitive index in the caecal (white bars) and colon content (grey bars) was analysed 5 days after inoculation (f; DSS, n = 5; DSS+W, n = 6). Data are shown as geometric mean and geometric s.d.
Supplementary information
Supplementary Table 1
This file contains information regarding the bacterial strains used in this study. (XLSX 11 kb)
Supplementary Table 2
This file contains information regarding primers used in this study. (XLSX 12 kb)
Supplementary Table 3
This file contains information regarding the plasmids used in this study. (XLSX 10 kb)
Supplementary Table 4
This file contains relevant information regarding the human samples used in this study. (XLSX 11 kb)
Supplementary Table 5
This file contains information regarding the statistical methods used as well as the details of statistics (such as exact p value) in each figure of the manuscript. (XLSX 28 kb)
Source data
Rights and permissions
About this article
Cite this article
Zhu, W., Winter, M., Byndloss, M. et al. Precision editing of the gut microbiota ameliorates colitis. Nature 553, 208–211 (2018). https://doi.org/10.1038/nature25172
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature25172
This article is cited by
-
The progression of inorganic nanoparticles and natural products for inflammatory bowel disease
Journal of Nanobiotechnology (2024)
-
Genetic evidence strengthens the bidirectional connection between gut microbiota and periodontitis: insights from a two-sample Mendelian randomization study
Journal of Translational Medicine (2023)
-
Sialic acid exacerbates gut dysbiosis-associated mastitis through the microbiota-gut-mammary axis by fueling gut microbiota disruption
Microbiome (2023)
-
Alternation of the gut microbiota in irritable bowel syndrome: an integrated analysis based on multicenter amplicon sequencing data
Journal of Translational Medicine (2023)
-
Dysbiosis of a microbiota–immune metasystem in critical illness is associated with nosocomial infections
Nature Medicine (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.