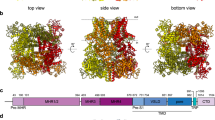
Abstract
TRPM4 is a calcium-activated, phosphatidylinositol-4,5-bisphosphate (PtdIns(4,5)P2) -modulated, non-selective cation channel that belongs to the family of melastatin-related transient receptor potential (TRPM) channels. Here we present the electron cryo-microscopy structures of the mouse TRPM4 channel with and without ATP. TRPM4 consists of multiple transmembrane and cytosolic domains, which assemble into a three-tiered architecture. The N-terminal nucleotide-binding domain and the C-terminal coiled-coil participate in the tetrameric assembly of the channel; ATP binds at the nucleotide-binding domain and inhibits channel activity. TRPM4 has an exceptionally wide filter but is only permeable to monovalent cations; filter residue Gln973 is essential in defining monovalent selectivity. The S1–S4 domain and the post-S6 TRP domain form the central gating apparatus that probably houses the Ca2+- and PtdIns(4,5)P2-binding sites. These structures provide an essential starting point for elucidating the complex gating mechanisms of TRPM4 and reveal the molecular architecture of the TRPM family.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
07 December 2017
In the HMTL, authors Xiao-chen Bai and Youxing Jiang were reversed in the author list.
References
Fleig, A. & Penner, R. The TRPM ion channel subfamily: molecular, biophysical and functional features. Trends Pharmacol. Sci. 25, 633–639 (2004)
Ramsey, I. S., Delling, M. & Clapham, D. E. An introduction to TRP channels. Annu. Rev. Physiol. 68, 619–647 (2006)
Venkatachalam, K. & Montell, C. TRP channels. Annu. Rev. Biochem. 76, 387–417 (2007)
Nadler, M. J. et al. LTRPC7 is a Mg·ATP-regulated divalent cation channel required for cell viability. Nature 411, 590–595 (2001)
Perraud, A. L. et al. ADP-ribose gating of the calcium-permeable LTRPC2 channel revealed by Nudix motif homology. Nature 411, 595–599 (2001)
Runnels, L. W., Yue, L. & Clapham, D. E. TRP-PLIK, a bifunctional protein with kinase and ion channel activities. Science 291, 1043–1047 (2001)
Guinamard, R., Demion, M. & Launay, P. Physiological roles of the TRPM4 channel extracted from background currents. Physiology (Bethesda) 25, 155–164 (2010)
Colquhoun, D., Neher, E., Reuter, H. & Stevens, C. F. Inward current channels activated by intracellular Ca in cultured cardiac cells. Nature 294, 752–754 (1981)
Launay, P. et al. TRPM4 is a Ca2+-activated nonselective cation channel mediating cell membrane depolarization. Cell 109, 397–407 (2002)
Nilius, B. et al. Voltage dependence of the Ca2+-activated cation channel TRPM4. J. Biol. Chem. 278, 30813–30820 (2003)
Pérez, C. A. et al. A transient receptor potential channel expressed in taste receptor cells. Nat. Neurosci. 5, 1169–1176 (2002)
Islam, M. S . (ed.) Transient Receptor Potential Channels, 147–171 (Springer Netherlands, 2011)
Zhang, Z., Okawa, H., Wang, Y. & Liman, E. R. Phosphatidylinositol 4,5-bisphosphate rescues TRPM4 channels from desensitization. J. Biol. Chem. 280, 39185–39192 (2005)
Nilius, B. et al. The Ca2+-activated cation channel TRPM4 is regulated by phosphatidylinositol 4,5-biphosphate. EMBO J. 25, 467–478 (2006)
Nilius, B., Prenen, J., Voets, T. & Droogmans, G. Intracellular nucleotides and polyamines inhibit the Ca2+-activated cation channel TRPM4b. Pflugers Arch. 448, 70–75 (2004)
Nilius, B. et al. Regulation of the Ca2+ sensitivity of the nonselective cation channel TRPM4. J. Biol. Chem. 280, 6423–6433 (2005)
Earley, S., Waldron, B. J. & Brayden, J. E. Critical role for transient receptor potential channel TRPM4 in myogenic constriction of cerebral arteries. Circ. Res. 95, 922–929 (2004)
Earley, S., Straub, S. V. & Brayden, J. E. Protein kinase C regulates vascular myogenic tone through activation of TRPM4. Am. J. Physiol. Heart Circ. Physiol. 292, H2613–H2622 (2007)
Del Negro, C. A. et al. Sodium and calcium current-mediated pacemaker neurons and respiratory rhythm generation. J. Neurosci. 25, 446–453 (2005)
Mironov, S. L. Metabotropic glutamate receptors activate dendritic calcium waves and TRPM channels which drive rhythmic respiratory patterns in mice. J. Physiol. (Lond.) 586, 2277–2291 (2008)
Cheng, H. et al. TRPM4 controls insulin secretion in pancreatic beta-cells. Cell Calcium 41, 51–61 (2007)
Launay, P. et al. TRPM4 regulates calcium oscillations after T cell activation. Science 306, 1374–1377 (2004)
Kruse, M. et al. Impaired endocytosis of the ion channel TRPM4 is associated with human progressive familial heart block type I. J. Clin. Invest. 119, 2737–2744 (2009)
Liu, H. et al. Gain-of-function mutations in TRPM4 cause autosomal dominant isolated cardiac conduction disease. Circ Cardiovasc Genet 3, 374–385 (2010)
Stallmeyer, B. et al. Mutational spectrum in the Ca2+-activated cation channel gene TRPM4 in patients with cardiac conductance disturbances. Hum. Mutat. 33, 109–117 (2012)
Liu, H. et al. Molecular genetics and functional anomalies in a series of 248 Brugada cases with 11 mutations in the TRPM4 channel. PLoS ONE 8, e54131 (2013)
Hof, T. et al. TRPM4 non-selective cation channel variants in long QT syndrome. BMC Med. Genet. 18, 31 (2017)
Rosenthal, P. B. & Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003)
Holm, L. & Rosenström, P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 (2010)
Kurakawa, T. et al. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445, 652–655 (2007)
Baugh, L. et al. Increasing the structural coverage of tuberculosis drug targets. Tuberculosis (Edinb.) 95, 142–148 (2015)
Jin, P. et al. Electron cryo-microscopy structure of the mechanotransduction channel NOMPC. Nature 547, 118–122 (2017)
Ullrich, N. D. et al. Comparison of functional properties of the Ca2+-activated cation channels TRPM4 and TRPM5 from mice. Cell Calcium 37, 267–278 (2005)
Demeuse, P., Penner, R. & Fleig, A. TRPM7 channel is regulated by magnesium nucleotides via its kinase domain. J. Gen. Physiol. 127, 421–434 (2006)
Zhang, Z. et al. The TRPM6 kinase domain determines the Mg·ATP sensitivity of TRPM7/M6 heteromeric ion channels. J. Biol. Chem. 289, 5217–5227 (2014)
Nightingale, E. R. Jr. Phenomenological theory of ion solvation. Effective radii of hydrated ions. J. Phys. Chem. 63, 1381–1387 (1959)
Nilius, B. et al. The selectivity filter of the cation channel TRPM4. J. Biol. Chem. 280, 22899–22906 (2005)
Long, S. B., Tao, X., Campbell, E. B. & MacKinnon, R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature 450, 376–382 (2007)
Payandeh, J., Scheuer, T., Zheng, N. & Catterall, W. A. The crystal structure of a voltage-gated sodium channel. Nature 475, 353–358 (2011)
Guo, J. et al. Structure of the voltage-gated two-pore channel TPC1 from Arabidopsis thaliana. Nature 531, 196–201 (2016)
Yamaguchi, S., Tanimoto, A., Otsuguro, K., Hibino, H. & Ito, S. Negatively charged amino acids near and in transient receptor potential (TRP) domain of TRPM4 channel are one determinant of its Ca2+ sensitivity. J. Biol. Chem. 289, 35265–35282 (2014)
Liao, M., Cao, E., Julius, D. & Cheng, Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature 504, 107–112 (2013)
Paulsen, C. E., Armache, J. P., Gao, Y., Cheng, Y. & Julius, D. Structure of the TRPA1 ion channel suggests regulatory mechanisms. Nature 520, 511–517 (2015)
Gao, Y., Cao, E., Julius, D. & Cheng, Y. TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature 534, 347–351 (2016)
Goehring, A. et al. Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat. Protoc. 9, 2574–2585 (2014)
Denisov, I. G., Grinkova, Y. V., Lazarides, A. A. & Sligar, S. G. Directed self-assembly of monodisperse phospholipid bilayer nanodiscs with controlled size. J. Am. Chem. Soc. 126, 3477–3487 (2004)
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017)
Zhang, K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016)
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012)
Bai, X. C., Rajendra, E., Yang, G., Shi, Y. & Scheres, S. H. Sampling the conformational space of the catalytic subunit of human γ-secretase. eLife 4, e11182 (2015)
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
DiMaio, F., Zhang, J., Chiu, W. & Baker, D. Cryo-EM model validation using independent map reconstructions. Protein Sci. 22, 865–868 (2013)
Amunts, A. et al. Structure of the yeast mitochondrial large ribosomal subunit. Science 343, 1485–1489 (2014)
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010)
Smart, O. S., Neduvelil, J. G., Wang, X., Wallace, B. A. & Sansom, M. S. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14, 354–360, 376 (1996)
Schrödinger, L. The PyMOL Molecular Graphics System, Version 1.8 (2015)
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004)
Acknowledgements
We thank N. Nguyen for manuscript preparation, J. M. Simard and J. Bryan for providing the TRPM4 clones. Single particle cryo-EM data were collected at the University of Texas Southwestern Medical Center (UTSW) Cryo-Electron Microscopy Facility. We thank D. Nicastro and Z. Chen for support in facility access and data acquisition. Negatively stained sample screening was performed at UTSW Electron Microscopy core. This work was supported in part by the Howard Hughes Medical Institute (Y.J.) and by grants from the National Institutes of Health (GM079179 to Y. J.) and the Welch Foundation (grant I-1578 to Y. J.). X.B. is supported by the Cancer Prevention and Research Initiative of Texas and Virginia Murchison Linthicum Scholar in Medical Research fund.
Author information
Authors and Affiliations
Contributions
J.G. and J.S. prepared the samples; J.G., J.S., Q.C. and X.B. performed data acquisition, image processing and structure determination; W.Z. performed electrophysiology; all authors participated in research design, data analysis, and manuscript preparation.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks R. Penner and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 Gating properties of mouse TRPM4 overexpressed in HEK293 cells.
a, Macroscopic currents of TRPM4 at ±100 mV in an inside-out patch in the presence or absence of various ligands in the bath (cytosolic). b, I–V curves of TRPM4 at the time points indicated in a. c, Sample traces of single channel recordings of TRPM4 in the Ca2+-desensitized state (with 300 μM Ca2+) at 100 mV (top), 0 mV (middle) and −100 mV (bottom), revealing the voltage-dependent, single-channel-open probability. d, Sample I–V curves of TRPM4 recorded with various cytosolic (bath solution) Ca2+ concentrations in the presence (right) or absence (left) of 10 μM PtdIns(4,5)P2. e, Concentration-dependent Ca2+-activation of TRPM4 channels in the presence or absence of PtdIns(4,5)P2. I/Imax values were measured at 100 mV from I–V curves shown in d. Data were reported as mean ± s.e.m. of five independent biological replicates. Curves are least-square fits to a Hill equation and the result indicates that Ca2+ has much lower apparent affinity for desensitized TRPM4. All electrophysiological recordings were repeated at least five times using different patches.
Extended Data Figure 2 Structure determination of apo TRPM4.
a, Purification of TRPM4 reconstituted in nanodiscs by size-exclusion chromatography. b, Negatively stained micrograph of TRPM4 in nanodiscs. c, Representative cryo-EM micrograph of TRPM4 in nanodiscs. d, Flowchart of image processing for apo TRPM4 particles. e, Gold-standard FSC curve of the final 3D reconstruction of the apo TRPM4 and the density map coloured by local resolution. f, Gold-standard FSC curve of particles from the focused 3D classification at the coiled-coil region and the density map coloured by local resolution.
Extended Data Figure 3 Structure determination of ATP-bound TRPM4.
a, Flow chart of image processing for ATP-bound TRPM4 particles. b, Gold-standard FSC curve of the final 3D reconstruction of the TRPM4–ATP complex and the density map coloured by local resolution. c, Gold-standard FSC curve of particles from the focused 3D classification at the coiled-coil region of TRPM4–ATP complex and the density map coloured by local resolution.
Extended Data Figure 4 Data collection, structure refinement and model validation.
a, Data collection and model refinement statistics. b, FSC curves for cross-validation between the maps and the models. Curves for model versus summed map in green (sum), for model versus half map in black (work), and for model versus half map not used for refinement in red (free).
Extended Data Figure 5 Sample EM density maps of TRPM4.
a, Sample maps at various regions of apo TRPM4. The maps are low-pass filtered to 3.1 Å and sharpened with a temperature factor of −120 Å2. b, EM density of coiled-coil region in the apo TRPM4 after focused 3D classification. The map is low-pass filtered to 3.5 Å and sharpened with a temperature factor of −91 Å2. c, Electron microscopy density of ATP and its surrounding residues in the ATP-bound TRPM4. The map is low-pass filtered to 2.9 Å and sharpened with a temperature factor of −91 Å2.
Extended Data Figure 6 Sequence alignment of mouse TRPM4 (MmTRPM4) and human TRPM (HsTRPM1–8) channels.
Secondary structure assignments are based on the mouse TRPM4 structure. Only the sequences up to the end of the coiled-coil domain are included in the alignment. Red triangles mark the key residues for ATP binding.
Extended Data Figure 7 NBD structure and ATP inhibition.
a, Structural comparison between the NBD of ATP-bound TRPM4 and AMP-bound LOG protein (PDB accession number 3SBX). The NBD region in the dotted box shares a similar fold to that of LOG protein. The ATP-binding site in the TRPM4 NBD is distinct from AMP in LOG. b, Superposition of TRPM4 structures in the apo (green) and ATP-bound (purple) states. The top two tiers are virtually identical in both states. Major conformational change occurs at the NBD. c. Sample I–V curves of TRPM4 and its mutants at various concentrations of cytosolic free ATP. Channels were activated by 300 μM Ca2+ and 10 μM PtdIns(4,5)P2 in the bath solution. Normalized currents (I/Imax) at 100 mV were used to generate the inhibition curves shown in Fig. 2e. Imax is the current at 100 mV without ATP. d, Recovery of wild-type TRPM4 and H160A mutant activities from Ca2+-desensitization by cytosolic ATP·Mg. Currents were recorded at −100 mV. Note that ATP was washed out before Ca2+ activation to avoid ATP inhibition. All electrophysiological recordings were repeated at least five times using different patches.
Extended Data Figure 8 The middle-tier LHD mediates inter-domain interactions within the subunit.
a, Structure of the LHD (cyan) and its interactions with the ARD (lower right inset), the TRP domain (upper right inset) and S1 helix (left inset). Between the middle and bottom tiers, the linker helices of LH1, LH4 and LH5 stack on top of AKR2 and form extensive hydrophobic interactions (lower right inset). The C-terminal region of the LHD mediates direct contacts with the top-tier transmembrane domain: the U-shaped LH9–LH11 grip the bottom side of TRP helix 1 (upper right inset); LH12 and the loops on its two ends clamp around the S0 and N terminus of S1 (left inset). b, Four linker domains encircle a wide open, fenestrated court at the centre of the channel. LH6–LH8 helices from each subunit frame the open central court and form head-to-tail packing with their neighbouring counterparts in a channel tetramer, providing the only inter-subunit contact at the middle tier.
Extended Data Figure 9 The Q973D mutant remains non-selective among monovalent cations similar to the wild-type TRPM4 channel.
Shown are sample I–V curves recorded in bi-ionic conditions with 150 mM Na+ in the pipette and 150 mM X+ (X = Li, Na, K or Cs) in the bath. Currents were recorded when channels reached desensitized steady state after activation with 300 μM cytosolic (bath) Ca2+. All recordings were repeated at least five times using different patches.
Extended Data Figure 10 S1–S4 domain of TRPM4.
a, Structural comparison of the S1–S4 domain between TRPM4 (green) and TRPV1 (blue, PDB accession number 5IRX). b, Voltage dependence of wild-type TRPM4 and R901A mutant at Ca2+-desensitized state (top). The membrane was stepped from the holding potential (0 mV) to various testing potentials (−100 mV to +100 mV) for one second and then stepped to −100 mV. The peak tail currents were used to plot the G–V curves (bottom). Data were reported as mean ± s.e.m. from five independent patches (biological replicates). Source Data are available in with the online version of the paper. V1/2 and Z values were obtained from fits of the data to the Boltzmann equation.
Supplementary information
Rights and permissions
About this article
Cite this article
Guo, J., She, J., Zeng, W. et al. Structures of the calcium-activated, non-selective cation channel TRPM4. Nature 552, 205–209 (2017). https://doi.org/10.1038/nature24997
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature24997
This article is cited by
-
Thermal gradient ring for analysis of temperature-dependent behaviors involving TRP channels in mice
The Journal of Physiological Sciences (2024)
-
TRPM channels in health and disease
Nature Reviews Nephrology (2024)
-
TRPM5 activation depends on a synergistic effect of calcium and PKC phosphorylation
Communications Biology (2024)
-
TRP (transient receptor potential) ion channel family: structures, biological functions and therapeutic interventions for diseases
Signal Transduction and Targeted Therapy (2023)
-
Structure of human TRPM8 channel
Communications Biology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.