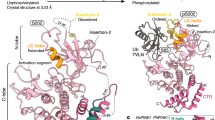
Abstract
Autosomal-recessive juvenile Parkinsonism (AR-JP) is caused by mutations in a number of PARK genes, in particular the genes encoding the E3 ubiquitin ligase Parkin (PARK2, also known as PRKN) and its upstream protein kinase PINK1 (also known as PARK6). PINK1 phosphorylates both ubiquitin and the ubiquitin-like domain of Parkin on structurally protected Ser65 residues, triggering mitophagy. Here we report a crystal structure of a nanobody-stabilized complex containing Pediculus humanus corporis (Ph)PINK1 bound to ubiquitin in the ‘C-terminally retracted’ (Ub-CR) conformation. The structure reveals many peculiarities of PINK1, including the architecture of the C-terminal region, and reveals how the N lobe of PINK1 binds ubiquitin via a unique insertion. The flexible Ser65 loop in the Ub-CR conformation contacts the activation segment, facilitating placement of Ser65 in a phosphate-accepting position. The structure also explains how autophosphorylation in the N lobe stabilizes structurally and functionally important insertions, and reveals the molecular basis of AR-JP-causing mutations, some of which disrupt ubiquitin binding.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Hoehn, M. M. & Yahr, M. D. Parkinsonism: onset, progression and mortality. Neurology 17, 427–442 (1967)
Corti, O., Lesage, S. & Brice, A. What genetics tells us about the causes and mechanisms of Parkinson’s disease. Physiol. Rev. 91, 1161–1218 (2011)
Valente, E. M. et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science 304, 1158–1160 (2004)
Clark, I. E. et al. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature 441, 1162–1166 (2006)
Park, J. et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature 441, 1157–1161 (2006)
Yang, Y. et al. Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc. Natl Acad. Sci. USA 103, 10793–10798 (2006)
Hewitt, V. L. & Whitworth, A. J. Mechanisms of Parkinson’s disease: lessons from Drosophila. Curr. Top. Dev. Biol. 121, 173–200 (2017)
Poole, A. C. et al. The PINK1/Parkin pathway regulates mitochondrial morphology. Proc. Natl Acad. Sci. USA 105, 1638–1643 (2008)
Pickrell, A. M. & Youle, R. J. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson’s disease. Neuron 85, 257–273 (2015)
Bingol, B. & Sheng, M. Mechanisms of mitophagy: PINK1, Parkin, USP30 and beyond. Free Radic. Biol. Med. 100, 210–222 (2016)
Kazlauskaite, A. & Muqit, M. M. K. PINK1 and Parkin – mitochondrial interplay between phosphorylation and ubiquitylation in Parkinson’s disease. FEBS J. 282, 215–223 (2015)
Narendra, D. P. et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 8, e1000298 (2010)
Okatsu, K. et al. PINK1 autophosphorylation upon membrane potential dissipation is essential for Parkin recruitment to damaged mitochondria. Nat. Commun. 3, 1016 (2012)
Okatsu, K. et al. A dimeric PINK1-containing complex on depolarized mitochondria stimulates Parkin recruitment. J. Biol. Chem. 288, 36372–36384 (2013)
Aerts, L., Craessaerts, K., De Strooper, B. & Morais, V. A. PINK1 kinase catalytic activity is regulated by phosphorylation on serines 228 and 402. J. Biol. Chem. 290, 2798–2811 (2015)
Kondapalli, C. et al. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol. 2, 120080 (2012)
Koyano, F. et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature 510, 162–166 (2014)
Kane, L. A. et al. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J. Cell Biol. 205, 143–153 (2014)
Kazlauskaite, A. et al. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem. J. 460, 127–139 (2014)
Wauer, T. et al. Ubiquitin Ser65 phosphorylation affects ubiquitin structure, chain assembly and hydrolysis. EMBO J. 34, 307–325 (2015)
Ordureau, A. et al. Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis. Mol. Cell 56, 360–375 (2014)
Wauer, T., Simicek, M., Schubert, A. & Komander, D. Mechanism of phospho-ubiquitin-induced PARKIN activation. Nature 524, 370–374 (2015)
Sauvé, V. et al. A Ubl/ubiquitin switch in the activation of Parkin. EMBO J. 34, 2492–2505 (2015)
Kumar, A. et al. Disruption of the autoinhibited state primes the E3 ligase parkin for activation and catalysis. EMBO J. 34, 2506–2521 (2015)
Kazlauskaite, A. et al. Binding to serine 65-phosphorylated ubiquitin primes Parkin for optimal PINK1-dependent phosphorylation and activation. EMBO Rep. 16, 939–954 (2015)
Sarraf, S. A. et al. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature 496, 372–376 (2013)
Lazarou, M. et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 524, 309–314 (2015)
Heo, J.-M., Ordureau, A., Paulo, J. A., Rinehart, J. & Harper, J. W. The PINK1-PARKIN mitochondrial ubiquitylation pathway drives a program of OPTN/NDP52 recruitment and TBK1 activation to promote mitophagy. Mol. Cell 60, 7–20 (2015)
Manning, G., Whyte, D. B., Martinez, R., Hunter, T. & Sudarsanam, S. The protein kinase complement of the human genome. Science 298, 1912–1934 (2002)
Woodroof, H. I. et al. Discovery of catalytically active orthologues of the Parkinson’s disease kinase PINK1: analysis of substrate specificity and impact of mutations. Open Biol. 1, 110012 (2011)
Gladkova, C. et al. An invisible ubiquitin conformation is required for efficient phosphorylation by PINK1. EMBO J. (2017). 10.15252/embj.201797876
Pleiner, T. et al. Nanobodies: site-specific labeling for super-resolution imaging, rapid epitope-mapping and native protein complex isolation. eLife 4, e11349 (2015)
Knighton, D. R. et al. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science 253, 407–414 (1991)
Lowe, E. D. et al. The crystal structure of a phosphorylase kinase peptide substrate complex: kinase substrate recognition. EMBO J. 16, 6646–6658 (1997)
Johnson, L. N., Noble, M. E. & Owen, D. J. Active and inactive protein kinases: structural basis for regulation. Cell 85, 149–158 (1996)
Taylor, S. S. & Kornev, A. P. Protein kinases: evolution of dynamic regulatory proteins. Trends Biochem. Sci. 36, 65–77 (2011)
Sim, C. H. et al. C-terminal truncation and Parkinson’s disease-associated mutations down-regulate the protein serine/threonine kinase activity of PTEN-induced kinase-1. Hum. Mol. Genet. 15, 3251–3262 (2006)
Gersch, M. et al. Mechanism and regulation of the Lys6-selective deubiquitinase USP30. Nat. Struct. Mol. Biol. 24, 920–930 (2017)
Aerts, L., De Strooper, B. & Morais, V. A. PINK1 activation-turning on a promiscuous kinase. Biochem. Soc. Trans. 43, 280–286 (2015)
Matenia, D., Hempp, C., Timm, T., Eikhof, A. & Mandelkow, E.-M. Microtubule affinity-regulating kinase 2 (MARK2) turns on phosphatase and tensin homolog (PTEN)-induced kinase 1 (PINK1) at Thr-313, a mutation site in Parkinson disease: effects on mitochondrial transport. J. Biol. Chem. 287, 8174–8186 (2012)
Dar, A. C., Dever, T. E. & Sicheri, F. Higher-order substrate recognition of eIF2α by the RNA-dependent protein kinase PKR. Cell 122, 887–900 (2005)
Kumar, A. et al. Structure of PINK1 and mechanisms of Parkinson’s disease associated mutations. eLife 6, 29985 (2017)
Hertz, N. T. et al. A neo-substrate that amplifies catalytic activity of Parkinson’s-disease-related kinase PINK1. Cell 154, 737–747 (2013)
Berrow, N. S. et al. A versatile ligation-independent cloning method suitable for high-throughput expression screening applications. Nucleic Acids Res. 35, e45 (2007)
Pardon, E. et al. A general protocol for the generation of nanobodies for structural biology. Nat. Protocols 9, 674–693 (2014)
Waterman, D. G. et al. Diffraction-geometry refinement in the DIALS framework. Acta Crystallogr. D Struct. Biol. 72, 558–575 (2016)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007)
Jiang, X. et al. Crystal structure of a LacY–nanobody complex in a periplasmic-open conformation. Proc. Natl Acad. Sci. USA 113, 12420–12425 (2016)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010)
Adams, P. D. et al. The Phenix software for automated determination of macromolecular structures. Methods 55, 94–106 (2011)
Silva, J. C. et al. Quantitative proteomic analysis by accurate mass retention time pairs. Anal. Chem. 77, 2187–2200 (2005)
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004)
Dong, X. et al. Ubiquitin S65 phosphorylation engenders a pH-sensitive conformational switch. Proc. Natl Acad. Sci. USA 114, 6770–6775 (2017)
Vallurupalli, P., Bouvignies, G. & Kay, L. E. Studying “invisible” excited protein states in slow exchange with a major state conformation. J. Am. Chem. Soc. 134, 8148–8161 (2012)
Holm, L. & Rosenström, P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 (2010)
Wen, Y. et al. Structural evaluation of a nanobody targeting complement receptor Vsig4 and its cross reactivity. Immunobiology 222, 807–813 (2017)
Vijay-Kumar, S., Bugg, C. E. & Cook, W. J. Structure of ubiquitin refined at 1.8 Å resolution. J. Mol. Biol. 194, 531–544 (1987)
Hamill, S., Lou, H. J., Turk, B. E. & Boggon, T. J. Structural basis for noncanonical substrate recognition of cofilin/ADF proteins by LIM kinases. Mol. Cell 62, 397–408 (2016)
Yokoyama, T., Kosaka, Y. & Mizuguchi, M. Structural insight into the interactions between death-associated protein kinase 1 and natural flavonoids. J. Med. Chem. 58, 7400–7408 (2015)
Wong, Y. L. et al. Reversible centriole depletion with an inhibitor of Polo-like kinase 4. Science 348, 1155–1160 (2015)
Pei, J., Kim, B.-H. & Grishin, N. V. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 36, 2295–2300 (2008)
Aguirre, J. D., Dunkerley, K. M., Mercier, P. & Shaw, G. S. Structure of phosphorylated UBL domain and insights into PINK1-orchestrated parkin activation. Proc. Natl Acad. Sci. USA 114, 298–303 (2017)
Acknowledgements
We thank beamline scientists at Diamond Light Source (DLS) for support at beamlines I04-1, I02 and I24; INSTRUCT, part of the European Strategy Forum on Research Infrastructures (ESFRI) and the Research Foundation - Flanders (FWO) for support with Nanobody discovery; A. Lundqvist for technical assistance during nanobody discovery; P. R. Elliott for help with crystallography and data collection; M. A. Michel, J. N. Pruneda, M. Gersch and other members of the D.K. laboratory for advice, reagents and discussions; and D. Barford and R. Williams for discussions and advice. Access to DLS was supported in part by the EU FP7 infrastructure grant BIOSTRUCT-X (contract 283570). The D.K. laboratory is supported by the Medical Research Council (U105192732), the European Research Council (309756, 724804), the Michael J. Fox Foundation and the Lister Institute for Preventive Medicine.
Author information
Authors and Affiliations
Contributions
A.F.S. and D.K. designed the study, and A.F.S. performed all experiments. C.G., S.M.V.F. and J.L.W. designed and characterized Ub-CR reagents, S.L.M. performed mass spectrometry, and E.P. and J.S. generated nanobodies. D.K. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
D.K. is part of the DUB Alliance, which includes Cancer Research Technology and FORMA Therapeutics.
Additional information
Reviewer Information Nature thanks J. W. Harper, T. Hunter and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 Similarity of PhPINK1 to other PINK1 orthologues.
PINK1 orthologues from different species were aligned using MUSCLE52. Secondary structure elements are shown and coloured as in Fig. 1. The β6 and the β9 sheets often observed in kinases form highly analogous interactions, but are not annotated as β-strand secondary structure elements in PhPINK1. The amino acid sequences of regions with no structural information are coloured grey. Catalytic residues and phosphorylation sites are highlighted in red and orange, respectively. The positions of insertions 1, 2, and 3 are highlighted in yellow. Stars indicate residues that are mutated in patients with AR-JP, and are coloured according to their structural roles (see Fig. 5 and Supplementary Table 1).
Extended Data Figure 2 Background of the Ub-CR conformation in Ub-TVLN.
a, When phosphorylated on Ser65, ubiquitin adopts an equilibrium between the common ubiquitin conformation and a second Ub-CR conformation, in which the β5 strand slides two residues further up its binding groove20,53. The panel illustrates the shape of ubiquitin and the relative positions of the β5 strand in the two conformations; a side view (boxed) shows how the hydrophobic Leu side chains slide in their respective pockets. b, Using CEST (chemical exchange saturation transfer) NMR experiments54, we found that a very small (‘invisible’) population of unphosphorylated wild-type ubiquitin adopts a Ub-CR conformation31. c, We were able to stabilize the Ub-CR conformation in unphosphorylated ubiquitin by mutating Thr66 to Val, and Leu67 to Asn. The resulting Ub-TVLN mutant adopts the Ub-CR conformation in the unphosphorylated state, binds PhPINK1 with higher affinity via its exposed Ser65-containing loop, and is consequently an approximately 50-fold more efficient substrate for PhPINK131. Phosphorylated Ub-TVLN also adopts the Ub-CR conformation31. d, Important structural differences between the common conformation of wild-type ubiquitin and the Ub-CR conformation of Ub-TVLN. The latter extends the Ser65 loop, which becomes flexible and solvent-exposed, while the C terminus is shortened. Moreover, the important Ile44 hydrophobic patch becomes disjointed as Val70 and His68 are shifted on the ubiquitin surface in the Ub-CR conformation. The new conformation therefore has several unique features that can be exploited by binding partners. Indeed, the properties of Ub-CR as a superior substrate and the knowledge that wild-type ubiquitin exists in the Ub-CR conformation are consistent with the idea that PINK1 utilizes this conformation during phosphorylation31.
Extended Data Figure 3 Nb696 stabilizes the PhPINK1–Ub-TVLN complex.
a, Pull-down experiments with Ub-TVLN and PhPINK1 (residues 115–575), using Nb696 as bait (B). L, loading; E, eluate. Nb696 does not pull down Ub-TVLN alone, interacts with PhPINK1 only weakly, but stably binds PhPINK1 and Ub-TVLN when both are present. Experiments were performed as technical duplicates. b, Gel filtration profile of PhPINK1 (residues 115–575) alone (dashed line) and PhPINK1 (residues 115–575) with Nb696 and Ub-TVLN. c, Gel filtration fractions showing that PhPINK1 co-elutes with Nb696 and Ub-TVLN. A protein contaminant (*), potentially a truncation product of PhPINK1, co-elutes with the complex. b and c correspond to a representative example from an experiment performed as duplicates. d, Melting temperatures of PhPINK1 (residues 115–575) alone, PhPINK1–Ub-TVLN, PhPINK1–Nb696, and PhPINK1–Nb696–Ub-TVLN. When both Nb696 and Ub-TVLN are added, the melting temperature of PhPINK1 increases by around 7 °C, consistent with complex stabilization. The left panel shows representative melting curves for each sample. The panel on the right plots the derived values. Experiments were performed as nine technical replicates. The average melting temperatures are indicated. e, f, Further optimization of the PhPINK1 construct by removal of flexible N-terminal residues resulted in PhPINK1 (residues 143–575), which also formed a stable trimeric complex with Nb696 and Ub-TVLN. Gel filtration runs with identical compositions were performed eleven times, and a representative experiment is shown. f, Coomassie-stained SDS–PAGE of protein-containing fractions of the representative experiment shown in e. Fractions from three gel filtration runs have been analysed with identical results. g, PhPINK1 (residues 143–575) was expressed in E. coli and is autophosphorylated in the PhPINK1-Nb696-Ub-TVLN complex at six to nine sites (top left). Dephosphorylation of the complex with λ-phosphatase (λ-PP) results in a species that is homogeneously phosphorylated at three sites on PhPINK1 (top right). λ-Phosphatase treatment does not affect intact mass of either Nb696 or Ub-TVLN in MgATP-free conditions (bottom). A second PhPINK1 species with lower abundance, which is presumed to be acetylated, was also detected and has the same phosphorylation pattern as the more abundant species (*, top left). The dephosphorylated complex was used for crystallization. All experiments were performed as technical duplicates.
Extended Data Figure 4 Electron density and structural details.
a, Electron density of the PhPINK1–Nb696–Ub-TVLN complex. The structure of the complex is shown as in Fig. 1d and covered by a 2|Fo|–|Fc| electron density map contoured at 1σ. Detailed views (boxes) show the same electron density map for each component and for selected interfaces (the latter at 1.5σ). b, Comparison of Nb696 from the PhPINK1–Nb696–Ub-TVLN complex structure with the two most similar structures in PDB according to DALI analysis55 (http://ekhidna.biocenter.helsinki.fi/dali_server): nanobody TP377 (PDB: 5E0Q32, beige, r.m.s.d. 0.624 Å) and a nanobody targeting mVsig4 (PDB: 5IMK56, light blue, r.m.s.d. 0.460 Å). The right panel shows the superposition of the structures. As expected, the stable nanobody scaffold is highly similar apart from divergent variable regions (red). c, Ub-TVLN from the PhPINK1–Nb696–Ub-TVLN complex (red) is structurally highly similar to recent Ub-CR structures (for example, Ub-L67S, PDB: 5OXI31, green, r.m.s.d. 0.766 Å). The structure is also highly similar to wild-type ubiquitin (PDB: 1UBQ57, grey, r.m.s.d. 0.669 Å) despite slippage of the last β-strand. The right panel shows the superposition of the structures. d, The structure of the PhPINK1–Nb696–Ub-TVLN complex as in Fig. 1e, but showing Nb696. Nb696 interacts with the ATP-binding site, N lobe, C lobe and Ub-TVLN. Nb696 does not contact the N-lobe phosphorylation sites, but binds on the opposing kinase surface (see Fig. 2). ATP is modelled from the phosphorylase kinase superposition (see Extended Data Fig. 5). e, Structural detail of nanobody interfaces. Hydrogen bonds are shown as dotted lines.
Extended Data Figure 5 PhPINK1 structure comparison and structure-based sequence alignment.
a, We used DALI55 to find known structures with high similarity to the structure of PhPINK1 in the PhPINK1–Nb696–Ub-TVLN complex. LIM domain kinase (LIMK1, PDB: 5HVK58, DALI score 23.6,), and death-associated protein kinase 1 (DAPK1, PDB: 5AV359, DALI score 23.5) had the highest similarity scores, but there were more than 100 kinase structures with DALI scores higher than 23. We then used the isolated, divergent N lobe of PhPINK1 in a second search, revealing high similarity to Polo-like kinase 4 (PLK4, PDB: 4YUR60, DALI score 9.5). We compared the structures of PhPINK1 and phosphorylase kinase (PDB: 2PHK34, DALI score 8.3), because the phosphorylase kinase structure has been characterized in various states, including bound to a substrate peptide (see Fig. 3). Left, PhPINK1 structure (coloured as in Fig. 2); middle, phosphorylase kinase; right, superposition. b, Structure-based sequence alignment of the PhPINK1 kinase domain and phosphorylase kinase from a, using PROMALS3D61. The PhPINK1 secondary structure elements are displayed as in Extended Data Fig. 1. c, Direct comparison with phosphorylase kinase reveals hallmarks of an active kinase conformation in PhPINK1. Left, spine analysis showing aligned C and R spines within the kinase core. Middle, two insets show the corresponding residues under a transparent surface. Right, expanded view into the ATP-binding site. In PhPINK1, ATP is modelled from the phosphorylase kinase superposition. Key structural elements include a crucial Glu–Lys salt bridge involving the αC helix, the DFG motif in the ‘in’ position, and the HRD motif in an active conformation. Striking similarities in the conformation of all elements support the notion that PhPINK1 is in an active conformation. d, The CTR extends the kinase domain and forms a contiguous hydrophobic core with the kinase C lobe. CTR helices are shown in purple, the kinase C lobe is shown in pink, and hydrophobic Leu, Ile, Phe and Trp residues are shown in stick representation. e, ConSurf analysis (https://consurf.tau.ac.il/2016/) of PhPINK1 (left) and a close-up of the CTR (right) coloured according to level of conservation. The CTR shows no apparent conserved patches except a hydrophobic patch between αK and αL that originates from Leu-zipper interactions between helices.
Extended Data Figure 6 Ub-TVLN and Ser65-loop binding to PhPINK1.
a, View focusing on the Ala46–Gly47 hairpin of Ub-TVLN, which occupies a pocket on the PhPINK1 N lobe between the Gly-rich loop, Tyr198, and insertion 3. PhPINK1 is shown under a surface to highlight shape complementarity. b, Ub-TVLN bound to PhPINK1, showing the position of Lys residues. Several Lys residues display disordered side chains and were modelled in their most favourable rotamers. Lys6 and Lys48 are close to the interface with PhPINK1, which may prevent binding to ubiquitin moieties that are ubiquitinated at these sites (that is, internal ubiquitin molecules in a ubiquitin polymer). This is consistent with recent evidence that PhPINK1 phosphorylates Lys6 and Lys48 chains with a preference for the distal ubiquitin38. c, d, Direct comparison of Ub-TVLN and wild-type ubiquitin binding to PhPINK1. c, In Ub-TVLN, the Ub-CR conformation enables optimized contacts with Val70, and allows His68 to contact the activation segment. d, In wild-type ubiquitin, the Gly47 hairpin is identical to the one in Ub-TVLN, and can form contacts with the N lobe, but Tyr198 in PhPINK1 is juxtaposed with the larger and more hydrophilic His68, and Val70 is not involved in the interaction. Moreover, the Ser65 loop is unable to contact the activation segment. The difference in the ability of the Ser65 loops to partake in the interaction with PhPINK1 was seen in NMR studies31. e, f, HDX-MS studies of PhPINK1 bound to Nb696 and Ub-TVLN or wild-type ubiquitin (see Methods). Experiments were performed as technical triplicates. Differences in PhPINK1 deuterium uptake upon complex formation with either Nb696+Ub-TVLN or Nb696+Ub WT are shown for 3 s, 30 s, and 300 s timepoints. Mass differences are plotted on the middle amino acids of the respective peptides. The data revealed identical binding sites for ubiquitin and Ub-TVLN in the N lobe of PhPINK1 (pink, yellow boxes), consistent with the complex structure. Importantly, Ub-TVLN also binds to the C-lobe activation segment (orange box), whereas this interaction is not seen with wild-type ubiquitin. It is also apparent that Ub-TVLN protects PhPINK1-binding sites more efficiently, suggesting that the Ub-TVLN-containing complex is more stable (compare 300 s datasets, red). g, h, Structure of the Ubl domain of Parkin resembles wild-type ubiquitin (green, Parkin Ubl-R33Q crystal structure, PDB: 3B1L, unpublished). g, Conservation of residues at the PhPINK1 interface (including Ala46–Gly47) would suggest a similar PhPINK1-binding mode to wild-type ubiquitin. A Ub-CR conformer for the Parkin Ubl has, however, not been reported, and a superposition (h) shows that, like wild-type ubiquitin, the Parkin Ubl domain would be unlikely to be able to bind the activation segment. NMR analysis, however, suggests that the Parkin Ubl Ser65 loop displays higher structural flexibility62.
Extended Data Figure 7 PhPINK1 autophosphorylation.
a, Autophosphorylation states of indicated bacterially expressed PhPINK1 (residues 115–575) variants, all of which were purified identically. The top row, middle panel, shows that these states can be collapsed by λ-phosphatase treatment to a singly phosphorylated species in the absence of Nb696 (compare with Extended Data Fig. 3) The identified phosphorylation state of the PhPINK1 variants is depicted in Figs 3f and 4b. Some mutations abrogate kinase autophosphorylation, while others (such as Y198E) are fully phosphorylated. Experiments were performed as technical duplicates. b, Structure of PhPINK1 from the complex, showing that phospho-Thr305 resides in an exposed area of the C lobe, far from the ATP- or ubiquitin-binding sites. It is unclear why this site has been spared by λ-phosphatase treatment; one explanation is that the neighbouring Pro306 may impede λ-phosphatase binding. c, Thermal denaturation analysis of PhPINK1 with mutated N-lobe autophosphorylation sites (S202A/S204A) or constructs lacking the ability to coordinate phospho-Ser202 with insertion 3 (R282A/N283A). Both constructs are already impaired in stability (by 3–4 °C compared to wild-type PhPINK1), suggesting that phosphorylation stabilizes the N lobe. Moreover, addition of Nb696 and Ub-TVLN, which increases the stability of wild-type PhPINK1 (see Extended Data Fig. 3), has no effect on either mutant. This is further evidence that phosphorylation is required for ubiquitin binding. The left panel shows representative melting curves for each sample. The right panel plots the derived values. Experiments were performed as nine technical replicates. The average melting temperatures are indicated. d, One autophosphorylation site that might have been dephosphorylated with λ-phosphatase treatment is on Ser375 in the activation segment (corresponding to Ser402 in hPINK1). Indeed, while the exposed position of Ser375 explains how it may have been removed by λ-phosphatase, the conformation of the activation segment in PhPINK1 does not reveal how the reportedly crucial phosphorylation event on this site13,15 impacts kinase activity. Traditionally, activation-segment phosphorylation is coordinated by the HRD motif (residues 332–334 in PhPINK1 and 360–362 in hPINK1), yet in PhPINK1 the shortest side-chain distance between Arg333 and Ser375 is around 13 Å; instead, Arg333 is in close proximity to the backbone of Lys380 and to the side chain of Asp379 (which also binds ubiquitin His68) (see Fig. 3c, Extended Data Fig. 6c). The activation segment conformation is supported by the αEF–αF loop below, and by intra-loop interactions such as Arg374–Gln378 and Lys380–Tyr404. On the other hand, the activation segment may be flexible enough for phospho-Ser375 to fold back and bind Arg333. It is also possible that phosphorylation of Ser375 (Ser402) is transiently important for kinase activation, and then no longer essential for ubiquitin phosphorylation. A structure of phosphorylated PhPINK1 at this site will be required to understand the reported importance of activation segment phosphorylation for kinase activity.
Extended Data Figure 8 Additional insight into AR-JP mutations in PINK1.
a, Detailed view showing PINK1 mutations that are not conserved, or those in positions where a direct impact on PINK1 structure is not obvious. Notably, these mutations are all annotated as heterozygous, and may exist in conjunction with other mutations. b, PhPINK1 mutations affecting the protein fold listed in Fig. 5a are shown in blue. This view of the surface of PhPINK1 shows that the mutated residues affect the core of the protein and have little effect on the surface. c, Detailed view of the activation segment, depicting the cluster of mutations shown in Fig. 5, as well as the exposed autophosphorylation site Ser375 (Ser402 in hPINK1). d, Detailed view of mutations in insertion 3, including the well-studied T313M (Ser285 in PhPINK1).
Supplementary information
Supplementary Information
This file contains Supplementary Figure 1, the uncropped versions of all SDS-PAGE gels and Supplementary Table 1, collated AR-JP mutations and their predicted structural effects. (PDF 1166 kb)
Rights and permissions
About this article
Cite this article
Schubert, A., Gladkova, C., Pardon, E. et al. Structure of PINK1 in complex with its substrate ubiquitin. Nature 552, 51–56 (2017). https://doi.org/10.1038/nature24645
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature24645
This article is cited by
-
Identification and structural characterization of small molecule inhibitors of PINK1
Scientific Reports (2024)
-
Activation mechanism of PINK1
Nature (2022)
-
Structural and functional consequences of NEDD8 phosphorylation
Nature Communications (2021)
-
Systematic analysis of PINK1 variants of unknown significance shows intact mitophagy function for most variants
npj Parkinson's Disease (2021)
-
Mitophagy in carcinogenesis and cancer treatment
Discover Oncology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.