Abstract
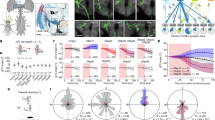
Nervous systems combine lower-level sensory signals to detect higher-order stimulus features critical to survival1,2,3, such as the visual looming motion created by an imminent collision or approaching predator4. Looming-sensitive neurons have been identified in diverse animal species5,6,7,8,9. Different large-scale visual features such as looming often share local cues, which means loom-detecting neurons face the challenge of rejecting confounding stimuli. Here we report the discovery of an ultra-selective looming detecting neuron, lobula plate/lobula columnar, type II (LPLC2)10 in Drosophila, and show how its selectivity is established by radial motion opponency. In the fly visual system, directionally selective small-field neurons called T4 and T5 form a spatial map in the lobula plate, where they each terminate in one of four retinotopic layers, such that each layer responds to motion in a different cardinal direction11,12,13. Single-cell anatomical analysis reveals that each arm of the LPLC2 cross-shaped primary dendrites ramifies in one of these layers and extends along that layer’s preferred motion direction. In vivo calcium imaging demonstrates that, as their shape predicts, individual LPLC2 neurons respond strongly to outward motion emanating from the centre of the neuron’s receptive field. Each dendritic arm also receives local inhibitory inputs directionally selective for inward motion opposing the excitation. This radial motion opponency generates a balance of excitation and inhibition that makes LPLC2 non-responsive to related patterns of motion such as contraction, wide-field rotation or luminance change. As a population, LPLC2 neurons densely cover visual space and terminate onto the giant fibre descending neurons, which drive the jump muscle motor neuron to trigger an escape take off. Our findings provide a mechanistic description of the selective feature detection that flies use to discern and escape looming threats.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Barlow, H. B. Summation and inhibition in the frog’s retina. J. Physiol. (Lond.) 119, 69–88 (1953)
Lettvin, J. Y ., Maturana, H. R ., McCulloch, W. S. & Pitts, W. H. What the frog’s eye tells the frog’s brain. Proc. Inst. Radio Eng. 47, 1940–1951 (1959)
Hubel, D. H. & Wiesel, T. N. Receptive fields, binocular interaction and functional architecture in the cat’s visual cortex. J. Physiol. (Lond.) 160, 106–154 (1962)
Gibson, J. J. Visually controlled locomotion and visual orientation in animals. Br. J. Psychol. 49, 182–194 (1958)
O’Shea, M. & Williams, J. The anatomy and output connection of a locust visual interneurone; the lobular giant movement detector (LGMD) neurone. J. Comp. Physiol. A 91, 257–266 (1974)
Wang, Y. & Frost, B. J. Time to collision is signalled by neurons in the nucleus rotundus of pigeons. Nature 356, 236–238 (1992)
de Vries, S. E. & Clandinin, T. R. Loom-sensitive neurons link computation to action in the Drosophila visual system. Curr. Biol. 22, 353–362 (2012)
Dunn, T. W. et al. Neural circuits underlying visually evoked escapes in larval zebrafish. Neuron 89, 613–628 (2016)
Peek, M. Y. & Card, G. M. Comparative approaches to escape. Curr. Opin. Neurobiol. 41, 167–173 (2016)
Wu, M. et al. Visual projection neurons in the Drosophila lobula link feature detection to distinct behavioral programs. eLife 5, e21022 (2016)
Maisak, M. S. et al. A directional tuning map of Drosophila elementary motion detectors. Nature 500, 212–216 (2013)
Takemura, S. Y. et al. A visual motion detection circuit suggested by Drosophila connectomics. Nature 500, 175–181 (2013)
Fischbach, K.-F. & Dittrich, A. The optic lobe of Drosophila melanogaster. I. A Golgi analysis of wild-type structure. Cell Tissue Res. 258, 441–475 (1989)
von Reyn, C. R. et al. A spike-timing mechanism for action selection. Nat. Neurosci. 17, 962–970 (2014)
Klapoetke, N. C. et al. Independent optical excitation of distinct neural populations. Nat. Methods 11, 338–346 (2014)
Nern, A., Pfeiffer, B. D. & Rubin, G. M. Optimized tools for multicolor stochastic labeling reveal diverse stereotyped cell arrangements in the fly visual system. Proc. Natl Acad. Sci. USA 112, E2967–E2976 (2015)
Schilling, T. & Borst, A. Local motion detectors are required for the computation of expansion flow-fields. Biol. Open 4, 1105–1108 (2015)
Seelig, J. D. et al. Two-photon calcium imaging from head-fixed Drosophila during optomotor walking behavior. Nat. Methods 7, 535–540 (2010)
Chen, T. W. et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300 (2013)
Strother, J. A. et al. The emergence of directional selectivity in the visual motion pathway of Drosophila. Neuron 94, 168–182 (2017)
Mauss, A. S. et al. Neural circuit to integrate opposing motions in the visual field. Cell 162, 351–362 (2015)
Fraser Rowell, C. H., O’Shea, M. & Williams, J. L. The neuronal basis of a sensory analyser, the acridid movement detector system. IV. The preference for small field stimuli. J. Exp. Biol. 68, 157–185 (1977)
Ishikane, H., Kawana, A. & Tachibana, M. Short- and long-range synchronous activities in dimming detectors of the frog retina. Vis. Neurosci. 16, 1001–1014 (1999)
Münch, T. A. et al. Approach sensitivity in the retina processed by a multifunctional neural circuit. Nat. Neurosci. 12, 1308–1316 (2009)
Gabbiani, F., Krapp, H. G., Koch, C. & Laurent, G. Multiplicative computation in a visual neuron sensitive to looming. Nature 420, 320–324 (2002)
Peron, S. & Gabbiani, F. Spike frequency adaptation mediates looming stimulus selectivity in a collision-detecting neuron. Nat. Neurosci. 12, 318–326 (2009)
Jones, P. W. & Gabbiani, F. Synchronized neural input shapes stimulus selectivity in a collision-detecting neuron. Curr. Biol. 20, 2052–2057 (2010)
Woolsey, T. A. & Van der Loos, H. The structural organization of layer IV in the somatosensory region (SI) of mouse cerebral cortex. The description of a cortical field composed of discrete cytoarchitectonic units. Brain Res. 17, 205–242 (1970)
von Reyn, C. R. et al. Feature integration drives probabilistic behavior in the Drosophila escape response. Neuron 94, 1190–1204 (2017)
Jenett, A. et al. A GAL4-driver line resource for Drosophila neurobiology. Cell Reports 2, 991–1001 (2012)
Kvon, E. Z. et al. Genome-scale functional characterization of Drosophila developmental enhancers in vivo. Nature 512, 91–95 (2014)
Pfeiffer, B. D. et al. Refinement of tools for targeted gene expression in Drosophila. Genetics 186, 735–755 (2010)
Hampel, S., Franconville, R., Simpson, J. H. & Seeds, A. M. A neural command circuit for grooming movement control. eLife 4, e08758 (2015)
Aso, Y. et al. The neuronal architecture of the mushroom body provides a logic for associative learning. eLife 3, e04577 (2014)
Wagh, D. A. et al. Bruchpilot, a protein with homology to ELKS/CAST, is required for structural integrity and function of synaptic active zones in Drosophila. Neuron 49, 833–844 (2006)
Iwai, Y. et al. Axon patterning requires DN-cadherin, a novel neuronal adhesion receptor, in the Drosophila embryonic CNS. Neuron 19, 77–89 (1997)
Peng, H., Ruan, Z., Long, F., Simpson, J. H. & Myers, E. W. V3D enables real-time 3D visualization and quantitative analysis of large-scale biological image data sets. Nat. Biotechnol. 28, 348–353 (2010)
Wan, Y., Otsuna, H., Chien, C. B. & Hansen, C. FluoRender: an application of 2D image space methods for 3D and 4D confocal microscopy data visualization in neurobiology research. In Proc. IEEE Pacific Visualization Symposium (eds Hauser, H., Kobourov, S. & Qu, H. ) 201–208 (IEEE, 2012)
Strother, J. A., Nern, A. & Reiser, M. B. Direct observation of ON and OFF pathways in the Drosophila visual system. Curr. Biol. 24, 976–983 (2014)
Franceschini, N. & Kirschfeld, K. Les phénoménes de pseudopupille dans l’oeil compose de Drosophila. Kybernetik 9, 159–182 (1971)
Pologruto, T. A., Sabatini, B. L. & Svoboda, K. ScanImage: flexible software for operating laser scanning microscopes. Biomed. Eng. Online 2, 13 (2003)
Wilson, R. I., Turner, G. C. & Laurent, G. Transformation of olfactory representations in the Drosophila antennal lobe. Science 303, 366–370 (2004)
Gabbiani, F., Krapp, H. G. & Laurent, G. Computation of object approach by a wide-field, motion-sensitive neuron. J. Neurosci. 19, 1122–1141 (1999)
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012)
Acknowledgements
We thank the Janelia FlyLight Project Team for help with brain dissections, histology, and confocal imaging, Janelia Scientific Computing for image processing and data analysis tools, the Janelia FlyLight Project Team, M. Wu, and T. Ngo for help with split-GAL4 screening, H. Dionne for split-GAL4 molecular biology, and E. Gruntman for feedback on stimulus design. We thank Janelia Instrument Design and Fabrication and Vidrio Technologies for advice and help with the two-photon microscope setup. We thank S. Namiki and the Janelia Descending Interneuron Project for the empty split-GAL4 line. We also thank K. Svoboda, A. Chuong, and members of the Card and Reiser laboratories for discussions and comments on the manuscript. This work was supported by the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
N.C.K., A.N., M.B.R. and G.M.C. designed all experiments. A.N. carried out all anatomical characterizations. M.Y.P. performed electrophysiology. P.B. performed behaviour experiments. A.N. and G.M.R. generated the split-GAL4 lines. E.M.R. generated additional combination lines. N.C.K. performed all calcium imaging. All authors contributed to the writing.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks T. Baden, H. Krapp and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 Additional LPLC2 anatomy.
a, Layer pattern of MCFO-labelled T4 and T5 neurons in the lobula plate. Individual cells arborize in one of the four lobula plate layers (arrows). A neuropil marker (anti-Brp) is shown in grey. The presynaptic terminals of T4/T5 in the lobula plate are mainly located in one of four Brp-rich strata. Scale bar, 10 μm. b–d, LPLC2 cells cover the lobula plate in overlapping patterns. b, As a population, LPLC2 dendrites (green) cover all lobula plate layers. Scale bar, 10 μm. c, Layer pattern for the LPLC2 cell shown in Fig. 1e, g on reference neuropil (grey, anti-Brp). LPLC2 arbors are mainly located in the Brp-rich layers that also contain the bulk of the presynaptic terminals of T4/T5 in the lobula plate (see a). Branches were manually coloured on the basis of layer position. d, MCFO-labelling of two LPLC2 cells in the lobula plate. Note the different positions of the layer 1 and layer 2 branches of the two cells. e, f, Additional examples of images of LPLC2 cells; images processed and displayed as in Fig. 1d, e (see Methods). Although details of the branching patterns vary, the general pattern of layer specific arbor spread appears stereotyped.
Extended Data Figure 2 Functional connectivity between T4/T5 and LPLC2.
A, B, Anatomy of fly transgenic used for functional connectivity experiments. Scale bar, 10 μm. A, Representative confocal image of Chrimson-expressing T4/T5 cells (red) and GCaMP6s-expressing LPLC2 cells (cyan) overlaid on neuropil marker (N-cadherin stain, grey). B, Two-photon averaged calcium image showing LPLC2 axon terminal imaging region. C, Experimental conditions for visual stimulus and optogenetic stimulation. Conditions a and b are looming at r/v = 40 ms, whereas conditions c and d have a static background intensity. D, E, Fly (N = 6) responses to visual and optogenetic stimuli. Individual fly responses are in grey and population average in black. Grey box indicates looming motion period. Red bar indicates red light stimulation period. E, Peak calcium response plotted for individual flies (circles) corresponding to measurements in D. F, G, Enhancerless GAL4 control fly (N = 6) responses to visual and optogenetic stimuli. H, Comparison of peak calcium responses to red light between empty (F, G) and T4/T5 (D, E) GAL4 driver lines. Individual fly responses (symbols) overlaid on population mean (line). Two-way ANOVA with Bonferroni’s post hoc test. *P < 0.05, ***P < 0.001.
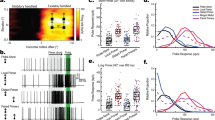
Extended Data Figure 3 Population LPLC2 looming selectivity and speed tuning.
A–E, LPLC2 axon terminal population calcium responses to dark looming and wide-field motion stimuli (N = 8 flies). Grey box on traces indicates stimulus motion period. A, B, Constant edge velocity looming (condition a, blue traces, 5–60° expansion, 10° s−1 edge speed) and wide-field motion stimuli (conditions b–g, black traces, 20° s−1 edge speed, 10° bar size in all applicable conditions). C–E, Constant edge velocity looming responses at the indicated speeds. Response during stimulus presentation is plotted either as peak fluorescence (D) or as instantaneous fluorescence as a function of disk diameter (E). D, Population mean ± 95% confidence intervals, with overlaid individual fly responses. E, Mean response (line) and 95% confidence intervals (shaded region). F, Looming-evoked escape rate under different effectors modulating LPLC2 activity or transmission. A total of 2,811 flies were assayed, with n > 130 flies in each condition shown (see Supplementary Table 3 for detailed statistics). Circle is jump rate; error bars, 95% confidence intervals. *P < 0.05 Tukey’s honest significant difference test.
Extended Data Figure 4 Single-cell LPLC2 receptive-field-centred responses.
A–L, LPLC2 single-axon responses to various dark/bright disk or ring stimuli (n = 10 neurons, N = 7 flies). All calcium traces shown depict individual neurons in grey and population average in black unless otherwise specified. Dashed grey circle in visual stimulus diagrams represents the measured 60° diameter upper limit of the LPLC2 receptive field. Overlaid grey box on calcium traces indicates motion stimulus period. Each neuron’s response is normalized to its dark looming response (a, 5–60° expansion). All dark and bright motion stimuli used constant edge velocity (10° s−1). Ring stimuli have fixed width of 5° (the difference between inner and outer radius). B, C, Individual neuron responses shown as circles. RM-ANOVA with Bonferroni’s post hoc test. ****P < 0.0001. D, E, Individual trial traces in response to expansion stimulus (A, a) are shown in D, and in response to contraction stimulus (A, b) are shown in E. F, Dark versus bright ring expansion traces (population mean and 95% confidence intervals). G, H, Receptive field size mapping. G, Calcium responses (top) to various size disk expansions (bottom). H, Calcium transients during pre-expansion static disk display (circle) and during-expansion responses plotted as a function of disk diameter (population mean ± 95% confidence intervals). I–L, One-dimensional inward bar motion responses for 10° (I, J) and 60° bars (K, L). Traces shown in (I, K) and plotted data in (J, L) are population average (dark) and 95% confidence intervals (light). M, Power measured at projection screen surface for darkening stimulus (A, e). Each data point is averaged over a 5.55-ms bin, corresponding to a single projector frame time.
Extended Data Figure 5 Further decomposition of excitatory and inhibitory inputs to LPLC2.
Decomposition of motion along cardinal axes (A–O) or between cardinal axes (P–S) from the same neurons as Fig. 4 (n = 10 neurons, N = 7 flies). Calcium traces are population average (black) and individual neurons (grey). Statistics analysed using RM-ANOVA with specified post hoc test throughout. Population plots are mean and 95% confidence intervals. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. A–J, Calcium traces and statistics are matched across each row (that is, A–C, D–F, etc.). Statistics plots on the left (B, E, H, K, N) compare the linear sum of individual responses (a, b) with the measured combined response (c), with individual neurons shown as circles and Bonferroni’s post hoc test. Statistics plots on the right (C, F, I, L, O) show the effect of orthogonal axis darkening and inward motion, Dunnett’s post hoc test using reference c. P, Q, Effects of bar width on expansion between cardinal axes. Dunnett’s post hoc test, reference 10° bar response. R, S, Decomposing responses to motion between cardinal axes. S, Left: comparison of the linear sum of individual responses at the receptive field centre (a) and edges (e) versus the measured response to expansion in both areas (c). Individual neurons shown as circles. Bonferroni’s post hoc test. S, Right: comparison of motion at the edge versus centre of the receptive field. Dunnett’s post hoc test, reference a.
Extended Data Figure 6 LPi4-3 directional tuning.
a, LPi4-3 (green) expression pattern in the lobula plate and the approximate region shown by two-photon imaging (yellow box). Scale bar, 20 μm. b, Two-photon maximum z-projection (60 μm depth). Scale bar, 20 μm. c, Directional tuning with 1 Hz temporal frequency square-wave grating displayed within a 50° square aperture. Overlaid bars represent stimulus onset (grey) and period of motion (blue). Individual fly responses shown in dark grey and population average shown in black (N = 6 flies). d–i, Spatially and temporally filtered calcium images (median filtered and temporally binned by four volumes) in response to dark-edge motion along the directions indicated from one representative fly. Five other flies showed a similar response. All images are shown on the same intensity scale. Each frame in (d, f, h) spans approximately 700 ms. The time-collapsed maximum projection images (e, g, i) show that different LPi4-3 boutons respond to front-to-back (e) versus top-to-bottom (i) edge motion, indicating a difference in directional preference.
Extended Data Figure 7 Effect of LPi4-3 depolarization on LPLC2 visual response properties.
LPLC2 single-cell visual responses in transgenic flies expressing CsChrimson in LPi4-3 cells and GCaMP6f in LPLC2 cells (n = 10 neurons, N = 7 flies). Calcium traces shown for individual neurons (grey) and population average (black) throughout. Each neuron’s response is normalized to its dark looming response (C, a, 5–60° expansion at r/v = 40 ms) throughout the figure. Overlaid light grey bar on calcium traces indicates motion stimulus period. Red bar indicates red light stimulation period. A, Maximum intensity projection image of CsChrimson-expressing LPi4-3 cells (red) and GCaMP6f-expressing LPLC2 cells (cyan) overlaid on neuropil marker (grey, N-cadherin stain). Scale bar, 10 μm. B, Two-photon imaging region is restricted to LPLC2 axons in the lobula. Scale bar, 10 μm. C–S, Visual responses to receptive-field-centred stimuli. C, Stimulus diagram (top) and calcium response (bottom) for constant approach-velocity looming (a–d, r/v = 40 ms) and constant edge expansion (e–g, 10° s−1 edge speed). D, Raw peak calcium responses to stimulus (C, a) used for normalization are shown for each neuron. E, Individual trial responses to stimulus (C, a). F, Individual trial responses to stimulus (C, b). G–S, Comparison of visual responses with or without red light (660 nm) stimulation (see Methods). Directional tuning with a 10° bar expanding outwards at 20° s−1 along the indicated directions. RM-ANOVA with Bonferroni’s post hoc test. *P < 0.05, **P < 0.01, ****P < 0.0001. G, Polar plot summary (means ± 95% confidence intervals). Statistically significant data points (P < 0.05) are drawn as filled circles, insignificant data points are drawn as open circles. H–S, Detailed single-cell traces and comparisons. Individual neurons depicted as circles in statistical comparisons.
Extended Data Figure 8 Controls for LPi4-3 optogenetic modulation experiments.
Single-cell visual responses from control flies with an LPLC2-driven GCaMP6f and an enhancerless GAL4-driven CsChrimson (n = 6 neurons, N = 6 flies). Calcium traces shown for individual neurons (grey) and population average (black) throughout. Each neuron’s response is normalized to its dark looming response (B, a, 5–60° expansion at r/v = 40 ms) throughout the figure. Overlaid light grey bar indicates motion stimulus period. Red bar indicates red light stimulation period. A, Representative calcium image from LPLC2 axons in the lobula. Scale bar, 10 μm. B–F, Visual responses to receptive-field-centred stimuli. B, Stimulus diagram (top) and calcium response trace (bottom) for constant approach-velocity looming (a–e, r/v = 40 ms) and constant edge expansion (f–h, 10° s−1 edge speed). C, Raw peak calcium responses to stimulus used for normalization (B, a) are shown for individual neurons. D, Effect of red light on looming responses (P = 0.2237, paired t-test, two-sided). E, Individual trial responses to stimulus (B, a). F, Individual trial responses to stimulus (B, c). G–W, Comparison of visual responses with or without red light (660 nm) stimulation. Directional tuning with a 10° bar expanding outwards at 20° s−1 along the indicated directions. RM-ANOVA with Bonferroni’s post hoc test, *P < 0.05. G, Polar plot summary (means ± 95% confidence intervals). Statistically significant data points (P < 0.05) are drawn as filled circles, insignificant data points are drawn as open circles. H–W, Detailed single-cell traces and comparisons. Individual neurons depicted as circles in statistical comparisons.
Extended Data Figure 9 Size and layer pattern of LPLC2 cells and putative inputs to LPLC2 dendrites.
a–c, En face views of the lobula plate from posterior show the spread of MCFO-labelled LPLC2 (a), LPi 4-3 (b), and T4/T5 (c) cells. Note the much larger spread of LPi cells compared with T4/T5 cells. Scale bars, 20 μm. d–f, Lobula plate layer pattern of LPLC2 (d), LPi4-3 (e), and T4/T5 cells (f). As previously described21, LPi cells project between layers with opposing T4/T5 preferred direction. Scale bar in d, 10 μm. Since lobula plate depth is not uniform, images in d–f are shown at similar but slightly different scales to facilitate comparison of layer patterns between images. Anti-Brp is shown in grey.
Extended Data Figure 10 LPLC2 single-cell responses to null stimuli and looming motion.
a, Wide-field translational motion stimuli (20° s−1 edge speed, 10° bar size in all conditions except pure edge motion). Individual neurons in grey, population average in black (n = 10 neurons, N = 7 flies). b, Constant edge velocity (10° s−1) disk expansion (60–80°) at various distances between disk centre and receptive field centre (each blue circle represents a different neuron) (n = 40 neurons, N = 7 flies). c, Paired comparison for off-centred disk expansion (α > 5°, 60–80° disk diameter) versus on-centred disk expansion (α < 5°, 5–20° disk diameter). The off-centred disk expansion is from b; the on-centred disk expansion is the peak response from receptive field centre mapping (as depicted in Fig. 3b–d). Paired t-test, two-sided. ****P < 0.0001. d, e, Constant approach-velocity looming. Individual neurons in grey, population average in black. Tuning curve is population mean and 95% confidence intervals (n = 10 neurons, N = 7 flies). f, Representative traces from a single LPLC2 neuron in response to different types of dark-edge motion. Left: outward edge motion along a single cardinal direction (left) or 15° offset from the cardinal axis (right). Cardinal axis denoted with a single green line. Middle: dark-edge motion (left) and off-centre disk expansion (right) on the order of receptive field size. Right: receptive-field-centred disk expansions from an initial diameter of 5° to a final diameter of 20° (left) or 60° (right).
Supplementary information
Supplementary Tables
This file contains Supplementary Tables 1-3. (PDF 344 kb)
LPLC2 dendritic branching pattern in the lobula plate
Rotating view of a single LPLC2 neuron showing branching pattern (same cell as Fig. 1d,e) on reference neuropil (grey, anti-Brp). Dendrites in each lobula plate layer are colored differently. (MOV 4733 kb)
Rights and permissions
About this article
Cite this article
Klapoetke, N., Nern, A., Peek, M. et al. Ultra-selective looming detection from radial motion opponency. Nature 551, 237–241 (2017). https://doi.org/10.1038/nature24626
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature24626
This article is cited by
-
Multilevel visual motion opponency in Drosophila
Nature Neuroscience (2023)
-
Synaptic gradients transform object location to action
Nature (2023)
-
Different spectral sensitivities of ON- and OFF-motion pathways enhance the detection of approaching color objects in Drosophila
Nature Communications (2023)
-
Neural Circuit Mechanisms Involved in Animals’ Detection of and Response to Visual Threats
Neuroscience Bulletin (2023)
-
Optic flow based spatial vision in insects
Journal of Comparative Physiology A (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.