Abstract
DNA repair is essential to prevent the cytotoxic or mutagenic effects of various types of DNA lesions, which are sensed by distinct pathways to recruit repair factors specific to the damage type. Although biochemical mechanisms for repairing several forms of genomic insults are well understood, the upstream signalling pathways that trigger repair are established for only certain types of damage, such as double-stranded breaks and interstrand crosslinks1,2,3. Understanding the upstream signalling events that mediate recognition and repair of DNA alkylation damage is particularly important, since alkylation chemotherapy is one of the most widely used systemic modalities for cancer treatment and because environmental chemicals may trigger DNA alkylation4,5,6. Here we demonstrate that human cells have a previously unrecognized signalling mechanism for sensing damage induced by alkylation. We find that the alkylation repair complex ASCC (activating signal cointegrator complex)7 relocalizes to distinct nuclear foci specifically upon exposure of cells to alkylating agents. These foci associate with alkylated nucleotides, and coincide spatially with elongating RNA polymerase II and splicing components. Proper recruitment of the repair complex requires recognition of K63-linked polyubiquitin by the CUE (coupling of ubiquitin conjugation to ER degradation) domain of the subunit ASCC2. Loss of this subunit impedes alkylation adduct repair kinetics and increases sensitivity to alkylating agents, but not other forms of DNA damage. We identify RING finger protein 113A (RNF113A) as the E3 ligase responsible for upstream ubiquitin signalling in the ASCC pathway. Cells from patients with X-linked trichothiodystrophy, which harbour a mutation in RNF113A, are defective in ASCC foci formation and are hypersensitive to alkylating agents. Together, our work reveals a previously unrecognized ubiquitin-dependent pathway induced specifically to repair alkylation damage, shedding light on the molecular mechanism of X-linked trichothiodystrophy.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Jackson, S. P. & Durocher, D. Regulation of DNA damage responses by ubiquitin and SUMO. Mol. Cell 49, 795–807 (2013)
Sirbu, B. M. & Cortez, D. DNA damage response: three levels of DNA repair regulation. Cold Spring Harb. Perspect. Biol. 5, a012724 (2013)
Zhao, Y., Brickner, J. R., Majid, M. C. & Mosammaparast, N. Crosstalk between ubiquitin and other post-translational modifications on chromatin during double-strand break repair. Trends Cell Biol. 24, 426–434 (2014)
Drabløs, F. et al. Alkylation damage in DNA and RNA—repair mechanisms and medical significance. DNA Repair 3, 1389–1407 (2004)
Fu, D., Calvo, J. A. & Samson, L. D. Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat. Rev. Cancer 12, 104–120 (2012)
Sedgwick, B., Bates, P. A., Paik, J., Jacobs, S. C. & Lindahl, T. Repair of alkylated DNA: recent advances. DNA Repair 6, 429–442 (2007)
Dango, S. et al. DNA unwinding by ASCC3 helicase is coupled to ALKBH3-dependent DNA alkylation repair and cancer cell proliferation. Mol. Cell 44, 373–384 (2011)
Wick, W. & Platten, M. Understanding and targeting alkylator resistance in glioblastoma. Cancer Discov. 4, 1120–1122 (2014)
Jung, D. J. et al. Novel transcription coactivator complex containing activating signal cointegrator 1. Mol. Cell. Biol. 22, 5203–5211 (2002)
Komander, D. & Rape, M. The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 (2012)
Prag, G. et al. Mechanism of ubiquitin recognition by the CUE domain of Vps9p. Cell 113, 609–620 (2003)
Liu, S . et al. Promiscuous interactions of gp78 E3 ligase CUE domain with polyubiquitin chains. Structure 20, 2138–2150 (2012)
Unk, I. et al. Human SHPRH is a ubiquitin ligase for Mms2-Ubc13-dependent polyubiquitylation of proliferating cell nuclear antigen. Proc. Natl Acad. Sci. USA 103, 18107–18112 (2006)
Zhao, G. Y. et al. A critical role for the ubiquitin-conjugating enzyme Ubc13 in initiating homologous recombination. Mol. Cell 25, 663–675 (2007)
Thorslund, T. et al. Histone H1 couples initiation and amplification of ubiquitin signalling after DNA damage. Nature 527, 389–393 (2015)
Hegele, A. et al. Dynamic protein-protein interaction wiring of the human spliceosome. Mol. Cell 45, 567–580 (2012)
Corbett, M. A. et al. A novel X-linked trichothiodystrophy associated with a nonsense mutation in RNF113A. J. Med. Genet. 52, 269–274 (2015)
Nakabayashi, K. et al. Identification of C7orf11 (TTDN1) gene mutations and genetic heterogeneity in nonphotosensitive trichothiodystrophy. Am. J. Hum. Genet. 76, 510–516 (2005)
Sowa, M. E., Bennett, E. J., Gygi, S. P. & Harper, J. W. Defining the human deubiquitinating enzyme interaction landscape. Cell 138, 389–403 (2009)
Zhao, Y. et al. Noncanonical regulation of alkylation damage resistance by the OTUD4 deubiquitinase. EMBO J. 34, 1687–1703 (2015)
Eng, J. K., McCormack, A. L. & Yates, J. R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 (1994)
Acknowledgements
We thank B. Sleckman, G. Oltz, T. Stappenbeck, S. Virgin, and K. Murphy for their advice on this manuscript. J.R.B. and A.K.B. are supported by a Cell and Molecular Biology Training Grant (5T32GM007067-40), and J.R.B. is also supported by a Shawn Hu and Angela Zeng Student Scholarship. J.M.S. is supported by a Monsanto Graduate Program Fellowship. P.M.L. is supported by a fellowship from the American Cancer Society (PF-14-182-01-DMC). We thank the patients and their families, whose help and participation made this work possible. We acknowledge the Alvin J. Siteman Cancer Center at Washington University and Barnes-Jewish Hospital for the use of the GEiC Core. The Siteman Cancer Center is supported by a National Cancer Institute Cancer Center Support Grant (P30 CA091842; Eberlein, PI). This work was supported by the National Institutes of Health (R01 GM108648 to A.V., R01 GM109102 to C.W., and R01 CA193318 to N.M.), the Alvin Siteman Cancer Research Fund, the Siteman Investment Program (both to N.M.), and the Children’s Discovery Institute of St. Louis Children’s Hospital (MC-II-2015-453 to N.M.).
Author information
Authors and Affiliations
Contributions
J.R.B., J.M.S., C.O., M.E.S. and N.M. performed cellular and biochemical experiments. P.M.L. performed isothermal titration calorimetry experiments. C.B.V. and R.R. performed alkylated lesion quantification. M.C.M., A.K.B. and Y.Z. assisted with providing reagents and technical help. P.M.L. and E.B. performed the structural analysis. M.A.C., J.G. and M.F. provided X-TTD patient cells. J.J. performed DNA fibre analysis and was supervised by A.V. C.W. supervised P.M.L. and E.B. G.S. supervised C.B.V. and R.R. N.M. supervised the project and wrote the manuscript with J.R.B., with input from all other authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
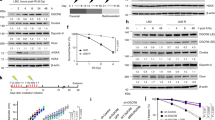
Extended Data Figure 1 The ASCC complex forms foci upon alkylation damage.
a, ASCC3 knockout cells were generated using CRISPR–Cas9 technology. Lysates were analysed by western blotting (n = 2 independent experiments). Clone 10 was verified to be a knockout by deep sequencing. b, Images of U2OS parental cells or ASCC3 knockout cells after MMS (n = 3 biological replicates). c, Immunofluorescence of U2OS cells after exposure to γ-irradiation (IR; 5 Gy) or ultraviolet (UV; 25 J m−2) (n = 3 biological replicates). d, Images of U2OS cells after treatment with the alkylating agents busulfan (4 mM), 1-(2-chloroethyl)-3-cyclohexyl-1-nitrosourea (CCNU; 100 μM), or temozolomide (TMZ; 1.0 mM) (n = 2 biological replicates). Numbers indicate the mean percentage of cells expressing five or more foci. e, Immunofluorescence of HA–ASCC2-expressing cells after exposure to the indicated damaging agents (n = 3 biological replicates). Scale bars, 10 μm. For gel source data, see Supplementary Fig. 1.
Extended Data Figure 2 Subcellular localization of ASCC2 and other alkylation repair factors.
a, Flow cytometry analysis of Flag–ASCC2-expressing cells after MMS treatment and Triton X-100 extraction. Numbers indicate the percentage of total cells in each quadrant (n = 2 independent experiments). b, Images of cells expressing HA–ASCC2 or HA–ALKBH3 after MMS treatment (n = 2 independent experiments). c, PLA quantification from Fig. 1c (n = 3 biological replicates; mean ± s.d.; two-tailed t-test, *P < 0.005). d, Immunofluorescence of cells expressing HA–ALKBH2, HA–methylguanine methyltransferase, or HA–alkyladenine glycosylase upon MMS treatment. e, Quantification of ASCC3 co-localization from d (n = 3 biological replicates; mean ± s.d.). Scale bars, 10 μm.
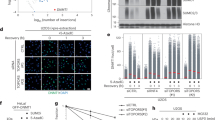
Extended Data Figure 3 Localization and interactions of the ASCC complex.
a, b, Images of U2OS or U2OS cells expressing the indicated vectors after MMS treatment (n = 3 biological replicates). c, Silver staining of the Flag–HA–ASCC2 complex purified from HeLa-S nuclear extract separated on 4–12% SDS–PAGE gel (n = 1 independent experiment). d, Tagged ASCC2 was purified with or without MMS and analysed by mass spectrometry. Peptide numbers for identified proteins were plotted for each condition. Expanded view is shown on the right (n = 1 independent experiment). e, f, Immunofluorescence analysis of U2OS or HA–ASCC2-expressing U2OS cells upon exposure to MMS (n = 3 biological replicates). g, U2OS cells were treated with MMS, and processed for immunofluorescence with or without initial incubation with RNase A (50 nM). Numbers indicate the percentage of cells expressing five or more ASCC3 foci (n = 3 biological replicates; mean ± s.d.). h, Biotinylated RNAs (20-mer, 35-mer, or 50-mer) were immobilized and tested for binding to recombinant His-NΔ-ASCC3 (n = 2 independent experiments). Scale bars, 10 μm.
Extended Data Figure 4 Functional interactions of the ASCC complex with other signalling pathways.
Immunofluorescence images of U2OS cells treated with MMS in the presence of spliceosomal inhibitor PLA-B (a) (100 nM; n = 3 biological replicates; mean ± s.d.), the RNA Pol II inhibitor DRB (b) (100 μM; n = 3 biological replicates; mean ± s.d.), or the indicated damage signalling kinase inhibitor (ATM 15 μM or ATR 10 μM) (c) (n = 2 biological replicates; mean). Numbers indicate the percentage of cells expressing five or more ASCC3 foci. d, Immunofluorescence of HA–ASCC2 and FK2 in cells after MMS (n = 3 biological replicates; mean ± s.d.). e, His-ASCC2 was purified on Ni-NTA, separated on a 10% SDS–PAGE gel, and analysed by Coomassie blue staining (n = 2 independent experiments). f, Immunofluorescence of HA–ASCC2 cells and K63-ubiquitin (top) or K48-ubiquitin (bottom) after MMS treatment (n = 2 independent experiments). Scale bars, 10 μm.
Extended Data Figure 5 ASCC2 binds specifically to K63-linked ubiquitin chains.
a, b, His-ASCC2 or the indicated His-ASCC2 deletions were immobilized on Ni-NTA and assessed for binding to K63-Ub2–7 (a) or K48-Ub2–7 (b) (n = 3 independent experiments). c, Schematic of ASCC2 or different ASCC2 deletions and their observed respective binding towards K63-Ub2–7 or K48-Ub2–7; n.d., not determined. d, Sequence alignment and conservation of residues 373–415 of human ASCC2. e, Interaction model between ubiquitin and the CUE domain of ASCC2 (PDB accession number 2DI0). The positions of four residues (L478, L479, P498, and L506) are shown. f, Binding assays were performed with K63-Ub2–7 using wild-type or the mutants of His-ASCC2 (n = 3 independent experiments).
Extended Data Figure 6 Characterization of ASCC2 knockout cells.
a, ASCC2 gene knockouts in U2OS and PC-3 cells were generated using CRISPR–Cas9 technology and verified by deep sequencing. Whole-cell lysates of the parental and knockout cells were analysed by western blotting as shown (n = 2 independent experiments). b, Flow cytometry of wild-type and ASCC2 knockout U2OS cells after MMS treatment to determine cell cycle distribution. c, Immunofluorescence analysis of HA–ALKBH2-expressing cells after MMS. Numbers indicate the percentage of cells expressing five or more HA–ALKBH2 foci (n = 3 biological replicates; mean ± s.d.). d, MMS sensitivity of wild-type or ASCC2 knockout cells using MTS assay (mean ± s.d.; n = 5 biological replicates). e, f, Sensitivity of wild-type and ASCC2 knockout cells to MMS (e) or camptothecin (f) was assessed by clonogenic survival assay (n = 4 biological replicates; mean ± s.d.). g, h, Wild-type PC-3 and ASCC2 knockout cell sensitivity to camptothecin (g) or bleomycin (h) using the MTS assay (n = 5 biological replicates; mean ± s.d.). i, Images of wild-type or ASCC2 knockout cells expressing the indicated vectors after MMS exposure. j, Quantification of i (n = 2 independent experiments; mean ± s.d.). k, Wild-type or ASCC2 knockout cells expressing indicated vectors were assessed for sensitivity to MMS using the MTS assay (n = 5 technical replicates; mean ± s.d.). Scale bar, 10 μm.
Extended Data Figure 7 ASCC2 coordinates ASCC–ALKBH3 complex recruitment during alkylation damage.
a, Whole-cell lysates from Extended Data Fig. 6i (left) and Fig. 3e (right) were collected and expression was analysed by western blotting (n = 2 independent experiments). b, Immunoprecipitation of HA–ASCC2 or HA–ASCC2 L506A was performed and analysed by western blot as shown (n = 2 biological replicates). c, Flag–ASCC2 or Flag–ALKBH3 was immobilized and tested for binding to full-length (FL) His-ASCC3 (n = 2 independent experiments). d, Flag–ASCC2 or Flag–ALKBH3 was immobilized and tested for binding to N-terminally deleted His-ASCC3 (His-ASCC3-ΔN) (n = 2 independent experiments). e, Flag–ALKBH3 was immobilized and tested for binding to His-ASCC2, with His-ASCC3-C (C terminus of ASCC3) serving as a positive control (n = 2 independent experiments). f, ASCC–ALKBH3 complex model.
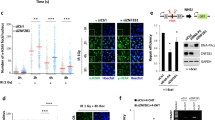
Extended Data Figure 8 Identification of the RNF113A E3 ligase.
a, Whole-cell lysates of U2OS cells infected with the indicated shRNAs were analysed by western blot. SHPRH was used as a loading control (n = 1 independent experiment). b, Immunofluorescence images of MMS-induced HA–ASCC2 foci in cells expressing the indicated shRNAs. c, HA–ASCC2 foci quantification from b (n = 3 biological replicates; mean ± s.d.; two-tailed t-test, *P < 0.001). d, Compilation of E3 ligase shRNA screen results. For each candidate, U2OS cells were transduced with HA–ASCC2 and an E3 targeting shRNA. MMS-induced HA–ASCC2 foci formation was analysed by immunofluorescence. Results were normalized to a scrambled shRNA (normalized score = 100). UBC13 denotes the positive control (purple). Results of three different shRNA to RNF113A are indicated in red (n = 1 independent experiment for each shRNA). e, Whole-cell lysates of U2OS cells infected with the indicated shRNAs were analysed by western blot. Asterisk indicates a non-specific band in the RNF113A blot (n = 2 independent experiments). f, Localization of Flag–ASCC2 and HA–RNF113A after MMS treatment (n = 3 biological replicates). g, Immunofluorescence of cells expressing Flag–RNF113A without MMS treatment (n = 3 biological replicates).
Extended Data Figure 9 Characterization of the E3 ubiquitin ligase activity of RNF113A and TTDN1.
a, TAP-RNF113A and the I264A RING finger mutant were stably expressed in HeLa-S cells and purified using anti-Flag resin. The eluted proteins were then analysed by silver staining after SDS–PAGE (n = 3 independent experiments). b, Ubiquitin ligase assays using E1, E2 (UbcH5c plus Ubc13/MMS2; 50 nM each), and wild-type or I264A RNF113A. Reactions were analysed by western blot (n = 3 independent experiments). c, MMS sensitivity of lymphoblasts from two patients with X-TTD compared with an unaffected individual (n = 5 biological replicates; mean ± s.d.). d, U2OS cells expressing the indicated combination of shRNA and RNF113A rescue vector were assessed for MMS sensitivity using an MTS assay (n = 5 technical replicates; mean ± s.d.). e, Whole-cell lysates of control or X-TTD lymphoblasts expressing indicated vectors after selection (n = 2 independent experiments). f, Immunofluorescence analysis of U2OS cells expressing the indicated shRNAs after MMS treatment. Western blot (n = 2 independent experiments) from the same cells is shown on the bottom, as is the quantification of ASCC3 foci (n = 3 biological replicates; mean ± s.d.).
Extended Data Figure 10 Functional characterization of RNF113A.
a, Schematic of human RNF113A and its domain structure. The three deletion constructs used for localization analysis are also shown. b, Images of cells expressing wild type or the indicated HA–RNF113A deletion constructs. Scale bar, 10 μm. Quantification of co-localization between each RNF113A construct and PRP8 is shown on the right (n = 3 biological replicates; mean ± s.d.). c, HEK293T cells expressing His-ubiquitin were transduced with control or RNF113A-targeting shRNAs and treated with MMS. Ubiquitinated proteins were isolated by Ni-NTA under denaturing conditions and western blotted as shown. Input lysates were also analysed as indicated. SF3B3, another ubiquitinated spliceosomal protein, was used as a control (n = 3 independent experiments). d, Cells expressing the indicated HA-vectors were treated with MMS as in c. Lysates were then used for ubiquitin pulldown assays with GST-ASCC2, then blotted as shown. Input lysates were also analysed as indicated (n = 2 independent experiments). e, His-NΔ-BRR2 was purified from Sf9 cells and analysed by SDS–PAGE and Coomassie staining (left). This was then used as a substrate for ubiquitination assays using HA–Ub and wild type or a RING-deletion (ΔRING) RNF113A (n = 2 independent experiments). f, Western blot analysis of U2OS cells expressing the indicated shRNAs used for immunofluorescence analysis in Fig. 4f (n = 2 independent experiments). g, Quantification of Fig. 4f (n = 3 biological replicates; mean ± s.d.; two-tailed t-test, #P < 0.001). h, MMS sensitivity of PC-3 cells expressing the indicated shRNAs was determined by MTS assay (n = 5 technical replicates; mean ± s.d.).
Supplementary information
Supplementary Figure 1
This file contains uncropped western blots and gels used in this study. Black rectangles denote how the blots and gels were cropped for final figures. (PDF 1851 kb)
Supplementary Table 1
Mass spectrometry data for TAP-ASCC2 purified from HeLa-S cells with or without prior exposure to MMS. (XLSX 138 kb)
Supplementary Table 2
A comprehensive list of each shRNA, including TRC numbers, used in the focused shRNA screen to identify the relevant E3 ligase. (XLSX 55 kb)
Supplementary Table 3
Mass spectrometry data for TAP-ASCC2 WT or the TAP-ASCC2 L506A CUE mutant purified from HeLa-S cells after MMS treatment. (XLSX 226 kb)
Supplementary Table 4
All antibodies used in this study with concentrations noted. The antibodies were produced in either rabbit or mouse. Applications include Western blot (WB), immunofluorescence microscopy (IF), flow cytometry (FC) and proximity ligation assay (PLA). (PDF 105 kb)
Rights and permissions
About this article
Cite this article
Brickner, J., Soll, J., Lombardi, P. et al. A ubiquitin-dependent signalling axis specific for ALKBH-mediated DNA dealkylation repair. Nature 551, 389–393 (2017). https://doi.org/10.1038/nature24484
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature24484
This article is cited by
-
Structural basis for clearing of ribosome collisions by the RQT complex
Nature Communications (2023)
-
Extended DNA threading through a dual-engine motor module of the activating signal co-integrator 1 complex
Nature Communications (2023)
-
ALKBH3 partner ASCC3 mediates P-body formation and selective clearance of MMS-induced 1-methyladenosine and 3-methylcytosine from mRNA
Journal of Translational Medicine (2021)
-
The role of ferroptosis in lung cancer
Biomarker Research (2021)
-
Demethyltransferase AlkBH1 substrate diversity and relationship to human diseases
Molecular Biology Reports (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.