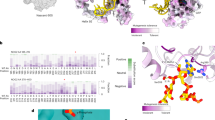
Abstract
Chemical modifications of human ribosomal RNA (rRNA) are introduced during biogenesis and have been implicated in the dysregulation of protein synthesis, as is found in cancer and other diseases. However, their role in this phenomenon is unknown. Here we visualize more than 130 individual rRNA modifications in the three-dimensional structure of the human ribosome, explaining their structural and functional roles. In addition to a small number of universally conserved sites, we identify many eukaryote- or human-specific modifications and unique sites that form an extended shell in comparison to bacterial ribosomes, and which stabilize the RNA. Several of the modifications are associated with the binding sites of three ribosome-targeting antibiotics, or are associated with degenerate states in cancer, such as keto alkylations on nucleotide bases reminiscent of specialized ribosomes. This high-resolution structure of the human 80S ribosome paves the way towards understanding the role of epigenetic rRNA modifications in human diseases and suggests new possibilities for designing selective inhibitors and therapeutic drugs.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Rodnina, M. V., Fischer, N., Maracci, C. & Stark, H. Ribosome dynamics during decoding. Phil. Trans. R. Soc. Lond. B 372, 1716 (2017)
Decatur, W. A. & Fournier, M. J. rRNA modifications and ribosome function. Trends Biochem. Sci. 27, 344–351 (2002)
Cantara, W. A. et al. The RNA Modification Database, RNAMDB: 2011 update. Nucleic Acids Res. 39, D195–D201 (2011)
Machnicka, M. A. et al. MODOMICS: a database of RNA modification pathways—2013 update. Nucleic Acids Res. 41, D262–D267 (2013)
Sharma, S. & Lafontaine, D. L. J. ‘View from a bridge’: a new perspective on eukaryotic rRNA base modification. Trends Biochem. Sci. 40, 560–575 (2015)
Maden, B. E. Identification of the locations of the methyl groups in 18 S ribosomal RNA from Xenopus laevis and man. J. Mol. Biol. 189, 681–699 (1986)
Maden, B. E. Locations of methyl groups in 28 S rRNA of Xenopus laevis and man: clustering in the conserved core of molecule. J. Mol. Biol. 201, 289–314 (1988)
Piekna-Przybylska, D., Decatur, W. A. & Fournier, M. J. The 3D rRNA modification maps database: with interactive tools for ribosome analysis. Nucleic Acids Res. 36, D178–D183 (2008)
Baxter-Roshek, J. L., Petrov, A. N. & Dinman, J. D. Optimization of ribosome structure and function by rRNA base modification. PLoS One 2, e174 (2007)
Ofengand, J. & Bakin, A. Mapping to nucleotide resolution of pseudouridine residues in large subunit ribosomal RNAs from representative eukaryotes, prokaryotes, archaebacteria, mitochondria and chloroplasts. J. Mol. Biol. 266, 246–268 (1997)
Green, R. & Noller, H. F. In vitro complementation analysis localizes 23S rRNA posttranscriptional modifications that are required for Escherichia coli 50S ribosomal subunit assembly and function. RNA 2, 1011–1021 (1996)
Auffinger, P. & Westhof, E. in Effects of Pseudouridylation on tRNA Hydration and Dynamics: A Theoretical Approach in Modification and Editing of RNA (eds Grosjean, H. & Benne, R. ) 103–112 (ASM, 1998)
Long, K. S. & Vester, B. Resistance to linezolid caused by modifications at its binding site on the ribosome. Antimicrob. Agents Chemother. 56, 603–612 (2012)
Benítez-Páez, A., Cárdenas-Brito, S., Corredor, M., Villarroya, M. & Armengod, M. E. Impairing methylations at ribosome RNA, a point mutation-dependent strategy for aminoglycoside resistance: the rsmG case. Biomedica 34 (Suppl. 1), 41–49 (2014)
Stojković, V., Noda-Garcia, L., Tawfik, D. S. & Fujimori, D. G. Antibiotic resistance evolved via inactivation of a ribosomal RNA methylating enzyme. Nucleic Acids Res. 44, 8897–8907 (2016)
Wirmer, J. & Westhof, E. Molecular contacts between antibiotics and the 30S ribosomal particle. Methods Enzymol. 415, 180–202 (2006)
Gonzales, B. et al. The Treacher Collins syndrome (TCOF1) gene product is involved in pre-rRNA methylation. Hum. Mol. Genet. 14, 2035–2043 (2005)
Freed, E. F., Bleichert, F., Dutca, L. M. & Baserga, S. J. When ribosomes go bad: diseases of ribosome biogenesis. Mol. Biosyst. 6, 481–493 (2010)
Penzo, M., Galbiati, A., Treré, D. & Montanaro, L. The importance of being (slightly) modified: the role of rRNA editing on gene expression control and its connections with cancer. Biochim. Biophys. Acta 1866, 330–338 (2016)
Lafontaine, D. L. J. Noncoding RNAs in eukaryotic ribosome biogenesis and function. Nat. Struct. Mol. Biol. 22, 11–19 (2015)
Bellodi, C. et al. H/ACA small RNA dysfunctions in disease reveal key roles for noncoding RNA modifications in hematopoietic stem cell differentiation. Cell Reports 3, 1493–1502 (2013)
Sloan, K. E. et al. Tuning the ribosome: The influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 2, 1–16 (2016)
Khatter, H., Myasnikov, A. G., Natchiar, S. K. & Klaholz, B. P. Structure of the human 80S ribosome. Nature 520, 640–645 (2015)
Myasnikov, A. G. et al. Structure–function insights reveal the human ribosome as a cancer target for antibiotics. Nat. Commun. 7, 12856 (2016)
Fischer, N. et al. Structure of the E. coli ribosome–EF-Tu complex at <3 Å resolution by Cs-corrected cryo-EM. Nature 520, 567–570 (2015)
Polikanov, Y. S., Melnikov, S. V., Söll, D. & Steitz, T. A. Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly. Nat. Struct. Mol. Biol. 22, 342–344 (2015)
Shalev-Benami, M. et al. 2.8-Å cryo-EM structure of the large ribosomal subunit from the eukaryotic parasite Leishmania. Cell Reports 16, 288–294 (2016)
von Loeffelholz, O. et al. Focused classification and refinement in high-resolution cryo-EM structural analysis of ribosome complexes. Curr. Opin. Struct. Biol. 46, 140–148 (2017)
Voorhees, R. M., Fernández, I. S., Scheres, S. H. W. & Hegde, R. S. Structure of the mammalian ribosome–Sec61 complex to 3.4 Å resolution. Cell 157, 1632–1643 (2014)
Limbach, P. A., Crain, P. F. & McCloskey, J. A. Summary: the modified nucleosides of RNA. Nucleic Acids Res. 22, 2183–2196 (1994)
Zorbas, C. et al. The human 18S rRNA base methyltransferases DIMT1L and WBSCR22-TRMT112 but not rRNA modification are required for ribosome biogenesis. Mol. Biol. Cell 26, 2080–2095 (2015)
Baudin-Baillieu, A. et al. Nucleotide modifications in three functionally important regions of the Saccharomyces cerevisiae ribosome affect translation accuracy. Nucleic Acids Res. 37, 7665–7677 (2009)
Liang, X.-H., Liu, Q. & Fournier, M. J. Loss of rRNA modifications in the decoding center of the ribosome impairs translation and strongly delays pre-rRNA processing. RNA 15, 1716–1728 (2009)
Meyer, B. et al. Ribosome biogenesis factor Tsr3 is the aminocarboxypropyl transferase responsible for 18S rRNA hypermodification in yeast and humans. Nucleic Acids Res. 44, 4304–4316 (2016)
Ito, S. et al. A single acetylation of 18 S rRNA is essential for biogenesis of the small ribosomal subunit in Saccharomyces cerevisiae. J. Biol. Chem. 289, 26201–26212 (2014)
Sharma, S. et al. Yeast Kre33 and human NAT10 are conserved 18S rRNA cytosine acetyltransferases that modify tRNAs assisted by the adaptor Tan1/THUMPD1. Nucleic Acids Res. 43, 2242–2258 (2015)
Hermann, T. & Westhof, E. Non-Watson–Crick base pairs in RNA–protein recognition. Chem. Biol. 6, R335–R343 (1999)
Swann, P. F. Why do O6-alkylguanine and O4-alkylthymine miscode? The relationship between the structure of DNA containing O6-alkylguanine and O4-alkylthymine and the mutagenic properties of these bases. Mutat. Res. 233, 81–94 (1990)
Sergeeva, O. V., Bogdanov, A. A. & Sergiev, P. V. What do we know about ribosomal RNA methylation in Escherichia coli? Biochimie 117, 110–118 (2015)
Melnikov, S. et al. One core, two shells: bacterial and eukaryotic ribosomes. Nat. Struct. Mol. Biol. 19, 560–567 (2012)
Hudson, B. H. & Zaher, H. S. O6-Methylguanosine leads to position-dependent effects on ribosome speed and fidelity. RNA 21, 1648–1659 (2015)
Roos, W. P., Thomas, A. D. & Kaina, B. DNA damage and the balance between survival and death in cancer biology. Nat. Rev. Cancer 16, 20–33 (2016)
Herzig, M. C. S. et al. DNA alkylating agent protects against spontaneous hepatocellular carcinoma regardless of O6-methylguanine-DNA methyltransferase status. Cancer Prev. Res. (Phila.) 9, 245–252 (2016)
Marcel, V . et al. p53 acts as a safeguard of translational control by regulating fibrillarin and rRNA methylation in cancer. Cancer Cell 24, 318–330 (2013)
Farley, K. I. & Baserga, S. J. Probing the mechanisms underlying human diseases in making ribosomes. Biochem. Soc. Trans. 44, 1035–1044 (2016)
Dinman, J. D. Pathways to specialized ribosomes: the Brussels lecture. J. Mol. Biol. 428 (10 Pt B), 2186–2194 (2016)
Slavov, N., Semrau, S., Airoldi, E., Budnik, B. & van Oudenaarden, A. Differential stoichiometry among core ribosomal proteins. Cell Reports 13, 865–873 (2015)
Preiss, T. All ribosomes are created equal. Really? Trends Biochem. Sci. 41, 121–123 (2016)
Marcel, V., Catez, F. & Diaz, J.-J. Ribosome heterogeneity in tumorigenesis: the rRNA point of view. Mol. Cell. Oncol. 2, e983755 (2015)
Krogh, N. et al. Profiling of 2′-O-Me in human rRNA reveals a subset of fractionally modified positions and provides evidence for ribosome heterogeneity. Nucleic Acids Res. 44, 7884–7895 (2016)
Khatter, H. et al. Purification, characterization and crystallization of the human 80S ribosome. Nucleic Acids Res. 42, e49 (2014)
Garreau de Loubresse, N. et al. Structural basis for the inhibition of the eukaryotic ribosome. Nature 513, 517–522 (2014)
Brodersen, D. E. et al. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell 103, 1143–1154 (2000)
Borovinskaya, M. A., Shoji, S., Fredrick, K. & Cate, J. H. D. Structural basis for hygromycin B inhibition of protein biosynthesis. RNA 14, 1590–1599 (2008)
Gürel, G., Blaha, G., Moore, P. B. & Steitz, T. A. U2504 determines the species specificity of the A-site cleft antibiotics: the structures of tiamulin, homoharringtonine, and bruceantin bound to the ribosome. J. Mol. Biol. 389, 146–156 (2009)
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017)
Zhang, K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016)
Scheres, S. H. W. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012)
Chen, S. et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy 135, 24–35 (2013)
Rosenthal, P. B. & Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003)
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014)
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010)
Natchiar, K. S., Myasnikov, A. G., Kratzat, H., Hazemann, I. & Klaholz, B. P. Atomic model building and refinement into high-resolution cryo-EM maps. Protocol Exchange http://dx.doi.org/10.1038/protex.2017.122 (2017)
Afonine, P. V. et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 (2012)
Lebedev, A. A. et al. JLigand: a graphical tool for the CCP4 template-restraint library. Acta Crystallogr. D Biol. Crystallogr. 68, 431–440 (2012)
Yang, H., Henning, D. & Valdez, B. C. Functional interaction between RNA helicase II/Guα and ribosomal protein L4. FEBS J. 272, 3788–3802 (2005)
Gigova, A., Duggimpudi, S., Pollex, T., Schaefer, M. & Koš, M. A cluster of methylations in the domain IV of 25S rRNA is required for ribosome stability. RNA 20, 1632–1644 (2014)
Sharma, S., Yang, J., Watzinger, P., Kötter, P. & Entian, K.-D. Yeast Nop2 and Rcm1 methylate C2870 and C2278 of the 25S rRNA, respectively. Nucleic Acids Res. 41, 9062–9076 (2013)
Schosserer, M. et al. Methylation of ribosomal RNA by NSUN5 is a conserved mechanism modulating organismal lifespan. Nat. Commun. 6, 6158 (2015)
Chow, C. S., Lamichhane, T. N. & Mahto, S. K. Expanding the nucleotide repertoire of the ribosome with post-transcriptional modifications. ACS Chem. Biol. 2, 610–619 (2007)
Acknowledgements
We thank J. Michalon, R. Fritz and R. David for IT support, J.-F. Ménétret for technical support, M.-C. Poterszman for constant support, the IGBMC cell culture facilities for HeLa cell production, and B. Beinsteiner for making the 3D animation. We thank D. Agard and S. Zheng for making MotionCor2 available ahead of publication. This work was supported by CNRS, Association pour la Recherche sur le Cancer (ARC), Institut National du Cancer (INCa), Ligue nationale contre le cancer (Ligue), Agence National pour la Recherche (ANR; ANR-10-LABX-0030-INRT under the program Investissements d’Avenir ANR-10-IDEX-0002-02). The electron microscope facility was supported by the Alsace Region, the Fondation pour la Recherche Médicale (FRM), Inserm, CNRS and ARC, by Instruct-ULTRA as part of the European Union’s Horizon 2020 (grant ID 731005), the French Infrastructure for Integrated Structural Biology (FRISBI; ANR-10-INSB-05-01) and by Instruct-ERIC.
Author information
Authors and Affiliations
Contributions
I.H. performed sample preparation, A.G.M. acquired cryo-EM data, A.G.M., H.K. and S.K.N. performed image processing, S.K.N. did structure refinement and model building, and S.K.N. and B.P.K. performed structural analysis of the rRNA. All authors analysed the data. B.P.K supervised the project and wrote the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks J. D. Dinman and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 Particle sorting scheme.
The initial dataset (top) was sorted into two main 3D classes (+/− rotated) and particles of the non-rotated state were either split further, depending on whether tRNA is bound to the E site (absence of tRNA means CHX is bound), or subjected to focused refinement of the 60S subunit and the 40S subunit head and body parts.
Extended Data Figure 2 Focused refinement and resolution estimation.
a, Focused refinement of the 60S subunit and the 40S subunit head and body regions (left, entire 80S complex; right, central section). b, Sections through the individually refined regions during focused refinement (the individually refined areas are sharp, whereas the other regions are less ordered). c, Individually refined regions in the 80S structure. d, Resolution estimation from the FSC curves.
Extended Data Figure 3 Representative regions in the 60S and 40S ribosomal subunits.
a–d, Cryo-EM map and atomic model of various regions in the 60S subunit. e–h, Cryo-EM map and atomic model of various regions in the 40S subunit.
Extended Data Figure 4 Register shift examples in previously less ordered rRNA regions.
a–f, Comparison of the previous map and previous atomic model23 (top), with the new map and the previous model (middle), and the new map with the refined atomic model after correction of register shifts.
Extended Data Figure 5 Specific features in the human ribosome structure.
a–c, Reannotation of an rRNA region as a ribosomal protein (eL29). d, Protein modifications on two lysine residues. e–h, Analysis of rRNA modifications in the 5.8S rRNA including sub-stoichiometric modification of Um14. i, j, Comparisons of neighbouring residues with and without rRNA modifications (human 60S and 40S ribosomal subunits, respectively).
Extended Data Figure 6 Annotation of chemical modifications in the 60S ribosomal subunit.
Conserved sites in E. coli and human large ribosomal subunits (magenta), predicted and found sites (cyan), unpredicted 2′-O-Me modification sites (blue), unpredicted base modification sites (red) and a 5.8S rRNA modification (green).
Extended Data Figure 7 Detailed views of the chemical modifications in the 60S ribosomal subunit (class I and class II).
Individual modification sites in classes I and II (magenta and cyan, respectively; cyan arrows indicate 2′-O-ribose methylations, black arrows indicate Ψs validated through the specific hydrogen-bond pattern, other modifications are indicated with magenta arrows).
Extended Data Figure 8 Detailed views of the chemical modifications in the 60S ribosomal subunit (class III).
Individual modification sites in class III (red; arrow colours as in Extended Data Fig. 7).
Extended Data Figure 9 Annotation of chemical modifications in the 40S ribosomal subunit.
Conserved sites in E. coli and human (magenta), predicted and found sites (cyan), unpredicted 2′-O-Me modification sites (blue) and unpredicted base modification sites (red).
Extended Data Figure 10 Detailed views of the chemical modifications in the 40S ribosomal subunit (classes I, II and III).
Individual modification sites in classes I, II and III (in magenta, cyan and red, respectively; arrow colours as in Extended Data Fig. 7).
Extended Data Figure 11 rRNA modifications in E. coli and human ribosomal subunits.
a, Large ribosomal subunit (left, E. coli; right, human). b, Small ribosomal subunit (left, E. coli; right, human).
Supplementary information
Supplementary Table 1
This file contains universally conserved sites (Class I) and bacteria-specific sites.
Supplementary Table 2
This file contains predicted sites (Class II) visible in the human ribosome structure and 2’-O-methylation (Am, Cm, Gm, Um); methylation at various at atomic positions (mn); pseudo-uridylation Ψ (not counting other predicted sites); aminocarboxypropylation (acp).
Supplementary Table 3
This file contains unpredicted human specific modifications (Class III) and a list sorted according to increasing residue numbers (Class III):
3D video of the human ribosome structure
The 3D video of the human ribosome structure shows the overall structure with the cryo-EM map (blue mesh) and the atomic model, then the full set of rRNA modifications and some more detailed examples of the 3 classes of rRNA modifications.
Rights and permissions
About this article
Cite this article
Natchiar, S., Myasnikov, A., Kratzat, H. et al. Visualization of chemical modifications in the human 80S ribosome structure. Nature 551, 472–477 (2017). https://doi.org/10.1038/nature24482
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature24482
This article is cited by
-
Methionine aminopeptidase 2 and its autoproteolysis product have different binding sites on the ribosome
Nature Communications (2024)
-
RNA modifications in physiology and disease: towards clinical applications
Nature Reviews Genetics (2024)
-
Heterogeneity of chemical modifications on RNA
Biophysical Reviews (2024)
-
Structural conservation of antibiotic interaction with ribosomes
Nature Structural & Molecular Biology (2023)
-
METTL5 serves as a diagnostic and prognostic biomarker in hepatocellular carcinoma by influencing the immune microenvironment
Scientific Reports (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.