Abstract
The skin barrier is the body’s first line of defence against environmental assaults, and is maintained by epithelial stem cells (EpSCs). Despite the vulnerability of EpSCs to inflammatory pressures, neither the primary response to inflammation nor its enduring consequences are well understood. Here we report a prolonged memory to acute inflammation that enables mouse EpSCs to hasten barrier restoration after subsequent tissue damage. This functional adaptation does not require skin-resident macrophages or T cells. Instead, EpSCs maintain chromosomal accessibility at key stress response genes that are activated by the primary stimulus. Upon a secondary challenge, genes governed by these domains are transcribed rapidly. Fuelling this memory is Aim2, which encodes an activator of the inflammasome. The absence of AIM2 or its downstream effectors, caspase-1 and interleukin-1β, erases the ability of EpSCs to recollect inflammation. Although EpSCs benefit from inflammatory tuning by heightening their responsiveness to subsequent stressors, this enhanced sensitivity probably increases their susceptibility to autoimmune and hyperproliferative disorders, including cancer.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
Change history
04 July 2018
In Fig. 2g of this Article, the 'D30 IMQ' image was inadvertently presented as a duplicate of the 'D6 Ctrl' image. The old, incorrect Fig. 2g is shown in the Supplementary Information to the accompanying Amendment. The original Article has been corrected (note scale bar of new panel).
References
Blanpain, C. & Fuchs, E. Stem cell plasticity. Plasticity of epithelial stem cells in tissue regeneration. Science 344, 1242281 (2014)
Alcolea, M. P. & Jones, P. H. Lineage analysis of epidermal stem cells. Cold Spring Harb. Perspect. Med. 4, a015206 (2014)
Adam, R. C. et al. Pioneer factors govern super-enhancer dynamics in stem cell plasticity and lineage choice. Nature 521, 366–370 (2015)
Hsu, Y.-C., Li, L. & Fuchs, E. Emerging interactions between skin stem cells and their niches. Nat. Med. 20, 847–856 (2014)
Kanneganti, T.-D. et al. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature 440, 233–236 (2006)
van der Fits, L. et al. Imiquimod-induced psoriasis-like skin inflammation in mice is mediated via the IL-23/IL-17 axis. J. Immunol. 182, 5836–5845 (2009)
Flutter, B. & Nestle, F. O. TLRs to cytokines: mechanistic insights from the imiquimod mouse model of psoriasis. Eur. J. Immunol. 43, 3138–3146 (2013)
Vasioukhin, V., Degenstein, L., Wise, B. & Fuchs, E. The magical touch: genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc. Natl Acad. Sci. USA 96, 8551–8556 (1999)
Li, M. et al. Topical vitamin D3 and low-calcemic analogs induce thymic stromal lymphopoietin in mouse keratinocytes and trigger an atopic dermatitis. Proc. Natl Acad. Sci. USA 103, 11736–11741 (2006)
Thomson, C. A., McColl, A., Cavanagh, J. & Graham, G. J. Peripheral inflammation is associated with remote global gene expression changes in the brain. J. Neuroinflammation 11, 73 (2014)
Levy, V., Lindon, C., Zheng, Y., Harfe, B. D. & Morgan, B. A. Epidermal stem cells arise from the hair follicle after wounding. FASEB J. 21, 1358–1366 (2007)
Naik, S. et al. Commensal-dendritic-cell interaction specifies a unique protective skin immune signature. Nature 520, 104–108 (2015)
Keyes, B. E. et al. Impaired epidermal to dendritic T cell signaling slows wound repair in aged skin. Cell 167, 1323–1338 (2016)
Dunn, L. et al. Murine model of wound healing. J. Vis. Exp. 75, 50265 (2013)
Naik, S. et al. Compartmentalized control of skin immunity by resident commensals. Science 337, 1115–1119 (2012)
Clark, R. A. Resident memory T cells in human health and disease. Sci. Transl Med. 7, 269rv1 (2015)
Netea, M. G. et al. Trained immunity: A program of innate immune memory in health and disease. Science 352, aaf1098 (2016)
Buenrostro, J. D., Wu, B., Chang, H. Y. & Greenleaf, W. J. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109, 21.29.1–21.29.9 (2015)
Feng, J., Liu, T., Qin, B., Zhang, Y. & Liu, X. S. Identifying ChIP-seq enrichment using MACS. Nat. Protocols 7, 1728–1740 (2012)
McLean, C. Y. et al. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501 (2010)
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010)
Beronja, S., Livshits, G., Williams, S. & Fuchs, E. Rapid functional dissection of genetic networks via tissue-specific transduction and RNAi in mouse embryos. Nat. Med. 16, 821–827 (2010)
DeYoung, K. L. et al. Cloning a novel member of the human interferon-inducible gene family associated with control of tumorigenicity in a model of human melanoma. Oncogene 15, 453–457 (1997)
Man, S. M., Karki, R. & Kanneganti, T.-D. AIM2 inflammasome in infection, cancer, and autoimmunity: role in DNA sensing, inflammation, and innate immunity. Eur. J. Immunol. 46, 269–280 (2016)
Dombrowski, Y. et al. Cytosolic DNA triggers inflammasome activation in keratinocytes in psoriatic lesions. Sci. Transl Med. 3, 82ra38 (2011)
Man, S. M. et al. Critical role for the DNA sensor AIM2 in stem cell proliferation and cancer. Cell 162, 45–58 (2015)
Wilson, J. E. et al. Inflammasome-independent role of AIM2 in suppressing colon tumorigenesis via DNA-PK and Akt. Nat. Med. 21, 906–913 (2015)
Hu, B. et al. The DNA-sensing AIM2 inflammasome controls radiation-induced cell death and tissue injury. Science 354, 765–768 (2016)
Zhang, F. et al. The caspase-1 inhibitor AC-YVAD-CMK attenuates acute gastric injury in mice: involvement of silencing NLRP3 inflammasome activities. Sci. Rep. 6, 24166 (2016)
Ge, Y. et al. Stem cell lineage infidelity drives wound repair and cancer. Cell 169, 636–650.e14 (2017)
Zhong, F. L. et al. Germline NLRP1 mutations cause skin inflammatory and cancer susceptibility syndromes via inflammasome activation. Cell 167, 187–202.e17 (2016)
Farshchian, M. et al. Tumor cell-specific AIM2 regulates growth and invasion of cutaneous squamous cell carcinoma. Oncotarget 8, 45825–45836 (2017)
Buch, T. et al. A Cre-inducible diphtheria toxin receptor mediates cell lineage ablation after toxin administration. Nat. Methods 2, 419–426 (2005)
Lochner, M. et al. In vivo equilibrium of proinflammatory IL-17+ and regulatory IL-10+ Foxp3+ RORγt+ T cells. J. Exp. Med. 205, 1381–1393 (2008)
Mainardi, S. et al. Identification of cancer initiating cells in K-Ras driven lung adenocarcinoma. Proc. Natl Acad. Sci. USA 111, 255–260 (2014)
Nguyen, H., Rendl, M. & Fuchs, E. Tcf3 governs stem cell features and represses cell fate determination in skin. Cell 127, 171–183 (2006)
Srinivas, S. et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1, 4 (2001)
Rabuffetti, M. et al. Inhibition of caspase-1-like activity by Ac-Tyr-Val-Ala-Asp-chloromethyl ketone induces long-lasting neuroprotection in cerebral ischemia through apoptosis reduction and decrease of proinflammatory cytokines. J. Neurosci. 20, 4398–4404 (2000)
Nowak, J. A. & Fuchs, E. Isolation and culture of epithelial stem cells. Methods Mol. Biol. 482, 215–232 (2009)
Rheinwald, J. G. & Green, H. Epidermal growth factor and the multiplication of cultured human epidermal keratinocytes. Nature 265, 421–424 (1977)
Dobin, A . et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013)
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014)
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. & Greenleaf, W. J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013)
Shin, H., Liu, T., Manrai, A. K. & Liu, X. S. CEAS: cis-regulatory element annotation system. Bioinformatics 25, 2605–2606 (2009)
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008)
Eisen, M. B., Spellman, P. T., Brown, P. O. & Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA 95, 14863–14868 (1998)
Speir, M. L. et al. The UCSC Genome Browser database: 2016 update. Nucleic Acids Res. 44, D717–D725 (2016)
Acknowledgements
We thank M. Nikolova, E. Wong and J. Levorse for technical assistance, and Y. Miao, I. Matos, Y. Ge, B. Keyes and R. Yi for discussions. FACS was conducted by Rockefeller’s Flow Cytometry Core (S. Mazel, director); ATAC-seq and RNA-seq were conducted by Rockefeller’s Genomics Core and Weill Cornell Genomics Center, respectively. E.F. is an Investigator of the Howard Hughes Medical Institute. S.N. is a Fellow supported by the Damon Runyon Cancer Research Foundation (DRG-2183-14) and L’Oreal USA For Women in Science. S.B.L. is funded by a National Institutes of Health (NIH) Ruth L. Kirschstein Predoctoral Fellowship (F31-AR068920-01A1). A.S. is funded by People Programme Marie Curie Actions (no. 629861). This study was supported by grants from the Robertson Foundation (S.N.), National Psoriasis Foundation (CEN5402062, S.N.), Pfizer (WI206828, S.N.) and NIH (R01-AR31737 and R01-AR050452, E.F.).
Author information
Authors and Affiliations
Contributions
S.N., S.B.L. and E.F. conceptualized the study, designed experiments and wrote the manuscript. S.N. and S.B.L. performed all animal, flow cytometry, microscopy, and genomic experiments. N.C.G. and A.S. analysed the ATAC–seq and RNA-seq datasets. K.A. performed qPCR validations, cell culture experiments and cloned the TRE-Aim2 overexpression construct. S.Y. assisted with culture studies and statistical analysis for wound healing studies. L.P. performed abrasion wound studies. A.K. helped to design the TRE-Aim2 overexpression construct. S.C. engineered the Krt10-creER mice.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks R. Flavell, X. Dai and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 Lineage tracing of skin stem cells and progeny during and after acute skin inflammation.
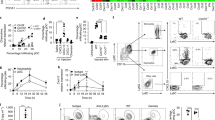
a, Epifluorescence images (left) and corresponding quantification (right) of TUNEL+ basal cells at day 6 of IMQ treatment (or vehicle control) and 30 days after treatment (n = 7; ***P < 0.0001). b, Schematic, immunofluorescence images and quantification of tamoxifen (TAM)-induced (corn oil control) RosaYFP reporter lineage tracing analysis with Krt14-creER (expressed by K14+ EpSCs) and Krt10-creER (expressed by K10+ terminally differentiating cells) (n = 4; all time points P > 0.05). Plots depict percentage of YFP+ cells relative to pre-IMQ (day 0) baselines (corresponding flow cytometric plots shown in c and e). Arrows denote YFP+ cells. c, Lineage tracing of Krt14-creER; RosaYFP at indicated times. Left, flow cytometric analysis of integrin α6+Sca1+CD34−YFP+ epidermal keratinocytes. Right, immunofluorescence of tamoxifen-activated EpSCs, lineage traced by YFP+ to include progeny (n = 3). d, Flow cytometry of Krt14-creER;Rosa YFP cells from the skin epidermis of animals that were lineage traced starting from IMQ treatment and analysed at day 180 (n = 2). e, Analysis of skin from Krt10-creER;RosaYFP mice that were lineage traced at the beginning of IMQ or control treatment (n = 3). Left, flow cytometric analysis of side YFP+ cells. Right, representative immunofluorescence images. Scale bars, 50 μm. Dotted lines demarcate the dermo-epidermal border. Arrows denote YFP+ keratinocytes. Data are mean ± s.e.m. n denotes the number of biologically independent animals per group. Significance for all plots was determined using two-tailed t-test at 95% confidence interval. All experiments have been replicated at least twice.
Extended Data Figure 2 Enhanced wound repair in post-inflamed epidermis.
a, One-phase decay modelling of wound repair in day 30 inflammation-experienced or vehicle-treated control mice (see Fig. 1b). Note, relative to two- or three-phase decay models (not shown), the data (shown at right) best fit the one-phase decay model, and this was therefore used for all subsequent wound repair data. b, Temporal wound closure analysis overlaid with one-phase decay analysis at day 180 after inflammation (n = 3). c, Temporal wound closure analysis overlaid with one-phase decay analysis of skin at day 30 after treatment with a variety of different inflammation-inducing agents: MC903, TPA, epidermal abrasion (wound), or fungal infection (C. albicans) (n = 4). See Fig. 1c for rate constants. d, Immunofluorescence images of wound edge labelled with the following antibodies: anti-EdU to mark proliferating cells, anti-K17 to mark wound-sensitized keratinocytes, integrin α5 to mark the migrating wound tongue that re-epithelializes the wound bed (w.b.), and K14, which marks the epidermal progenitors, expanded at the wound site. Vertical dotted lines denote the initial wound edge; arrows mark the edge of the extended epithelial tongue (n = 3). Scale bars, 100 μm. See Fig. 1e for quantifications. e, Representative images of silicone splinted 3 mm full thickness wounds from day 30 control and post-inflamed animals (n = 4). Scale bars, 3 mm. See Fig. 1f for quantifications. f, Migration assays were performed on skin explants (see Fig. 1g), in the presence or absence of mitomycin C for 5 days under conditions that quantitatively abrogate keratinocyte cell proliferation. Note that epidermal migration rates are similar, irrespective of whether cell proliferation was impaired (n = 3, 3 technical replicates per animal; P > 0.05, two-tailed t-test). Data are mean ± s.e.m. For individual data points in b and c, see Source Data. n denotes the number of biologically independent animals per group. All experiments have been replicated at least twice.
Extended Data Figure 3 The wound-healing advantage conferred to EpSCs is confined to the site of inflammation and occurs even when skin RORC+ cells are ablated.
a, Temporal wound closure analysis overlaid with one-phase decay analysis of inflammation-experienced and vehicle control skins, comparing the wound closure rates at sites distal and local to the topical application. Wound healing was initiated at day 30 after IMQ treatment, a time at which morphological signs of epidermal homeostasis were restored (n = 3). Plot display data combined from 3 independent experiments; see rate constants in Fig. 2b. b, c, Flow cytometric analysis of total immune cells (CD45+), αβ T cell receptor (TCR) T cells, dermal γδ T cells (γδ TCRlow), dendritic epidermal T cells (DETC; γδ TCRhigh), Langerhans cells (LC), dermal dendritic cells (DDC), macrophages (Mac) and eosinophils (Eos) from control and day 30 post-inflamed skins. Quantification shown at the right (n ≥ 3). d, Immunofluorescence images (left) and quantification (right) of T cells from Rorc-eGFP mice at the peak (day 6) of inflammation (see Fig. 2e for post-inflammation) (n = 3; ***P = 0.0056). Arrows denote RORC+CD3ε+ cells, thought to be the major drivers of IMQ-induced inflammation. Dotted lines demarcate the dermo-epidermal borders. e, Flow cytometric analysis of frequency and cellular distribution of RORC+ cells at day 30 post-inflammation (n = 4; ***P = 0.0005). The proportion of αβ TCR, γδ TCRlow and double-negative (DN; innate lymphoid) cells within the RORC(GFP)+ gate are shown adjacent to the quantification of total GFP+ cells. f, Depletion of RORC+ T cells does not result in a compensatory increase in other skin T cell populations (n = 2). g, Immunofluorescence of skin sections showing effective diphtheria toxin (DT)-mediated ablation of all CD3ε+RORC+ cells (yellow) in Rorc-DTR mice (n = 3), which activate DTR from the Rosa26 locus only in RORC+ cells. h, Wounds heal faster in post-inflamed skin despite ablation of skin RORC+ cells (n = 3). For corresponding rate constants in e–g, see Fig. 2f. i, Despite the absence of T and B cells, Rag2-null mice still mount a response to IMQ and display an accelerated wound-healing response after returning to homeostasis at day 30 post-inflammation (n = 3). For rate constants, see Fig. 2h. Scale bars, 100 μm (d) and 50 μm (g). Data are mean ± s.e.m. For individual data points in a, h and i, see Source Data. n denotes the number of biologically independent animals per group. Significance for all plots was determined using a two-tailed t-test at 95% confidence interval. All experiments have been replicated at least twice.
Extended Data Figure 4 Analysis of accessible chromatin in EpSCs during and after inflammation.
a, Immunofluorescence analysis of basal EpSC-specific markers in inflamed and control skin. Scale bar, 100 μm. b, FACS strategy for isolation of EpSCs (integrin α6+β1+Sca1+), with exclusion of CD45+, CD31+, CD117+ and CD140α+ non-epidermal cells, as well as dead (DAPI+), doublets (side-scatter width (SSC-Whigh) and forward-scatter width (FSC-Whigh)). c, qPCR validation of EpSC purity (left) using total mouse RNA as a control (right). Trp63 and Klf5 are specific for EpSCs; the others are not expressed by EpSCs (n = 3 mice pooled per group). d, Density plots depicting enrichment of ATAC–seq signals at TSSs ± 3 kb and around CTCF factor-binding sites. The x axis depicts respective distance ± 1 kb from each of these domains. e, Distribution of ATAC–seq peaks within defined genomic regions. UTR, untranslated regions of predicted mRNAs. f, Genomic browser shots of peaks enriched in EpSC-specific genes Klf5 and Krt14 and unaffected by IMQ. Arrows denote direction of transcription. g, Top, absolute numbers of ATAC–seq peaks from day 6 inflamed, day 30/180 post-inflamed and control EpSCs. Bottom, numbers and frequencies of ATAC–seq peaks that are shared in day 6 inflamed and either day 30 or day 180 post-inflamed EpSCs. h, Transcription factor motif enrichment (cumulative binomial distributions, P < 10−12) within the ATAC–seq peaks of day 6 inflamed EpSCs (n ≥ 3). For further details, see Fig. 3a–e. n denotes the number of biologically independent animals per group. All experiments have been replicated at least twice.
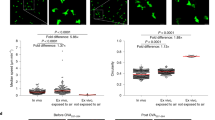
Extended Data Figure 5 Enrichment of inflammasome transcripts in wound-edge EpSCs of skin exposed to inflammation.
a, b, FACS strategy and qPCR verification of wound-edge EpSC purity from skin that was treated with either IMQ or vehicle and then allowed to return to homeostasis before wounding at day 30 (n = 3 pooled mice per group). Relevant antibodies and exclusion of dead cells and doublets are described in Extended Data Fig. 4. c, Matched ATAC–seq and RNA-seq analysis reveal that 91% of differentially expressed genes in day 6 inflamed versus control EpSCs are associated with newly acquired ATAC–seq peaks. For further data, see Fig. 4c–e. d, Sustained Aim2 transcription at day 30 post-inflammation in mice depleted of RORC+ cells (n = 3). For further data, see Fig. 4f. (Two-tailed t-test with 95% confidence interval, **P = 0.0028). e, Aim2-knockout mice do not show enhanced wound healing after inflammation (n = 3). For further data, see Fig. 4g. f, Aim2 induction in EpSCs is sufficient to augment wound healing in naive mice (n = 4). For further data, see Fig. 4h. For individual data points in e and f, see Source Data. Data are mean ± s.e.m. n denotes the number of biologically independent animals per group. Experiments in a–e have been replicated at least twice, experiment in f was performed once.
Extended Data Figure 6 Dissecting the AIM2 downstream effectors.
a, Model depicting possible effectors downstream of AIM2. b, c, AKT(Ser473) expression (b) and TUNEL labelling (c) in control and day 30 post-inflamed wounds (n = 3). d, The CASP1 inhibitor Ac-YVAD-cmk reduces re-epithelialization rates of wounded, post-inflamed skin compared with the rates of naive, vehicle control skin (n = 3). See Fig. 5b for schematic and further data. e, The absence of Il18 does not hamper the enhanced injury response of post-inflamed skin (n = 3). For further data, see Fig. 5d. f, The absence of IL-1β signalling, achieved in Il1r1-null mice, abrogates the enhanced injury response of post-inflamed skin (n = 4). For further data, see Fig. 5e. g, Anti-IL-1R1 treatment reverses the wound repair advantage conferred by epidermal Aim2 overexpression in naive mice (n = 4). For further data, see Fig. 5g. For individual data points in d–g, see Source Data. Scale bars, 100 μm (b) and 50 μm (c). n = 3. Data are mean ± s.e.m. n denotes the number of biologically independent animals per group. Experiments in a–f have been replicated at least twice, experiment in g was performed once.
Supplementary information
Source data
Rights and permissions
About this article
Cite this article
Naik, S., Larsen, S., Gomez, N. et al. Inflammatory memory sensitizes skin epithelial stem cells to tissue damage. Nature 550, 475–480 (2017). https://doi.org/10.1038/nature24271
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature24271
This article is cited by
-
Hypoxia inducible factor-1α regulates microglial innate immune memory and the pathology of Parkinson’s disease
Journal of Neuroinflammation (2024)
-
Sexual dimorphism in melanocyte stem cell behavior reveals combinational therapeutic strategies for cutaneous repigmentation
Nature Communications (2024)
-
Memory B cell subsets have divergent developmental origins that are coupled to distinct imprinted epigenetic states
Nature Immunology (2024)
-
Cellular and molecular mechanisms of skin wound healing
Nature Reviews Molecular Cell Biology (2024)
-
Beyond genetics: driving cancer with the tumour microenvironment behind the wheel
Nature Reviews Cancer (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.