Abstract
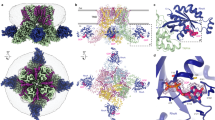
Transient receptor potential mucolipin 1 (TRPML1) is a Ca2+-releasing cation channel that mediates the calcium signalling and homeostasis of lysosomes. Mutations in TRPML1 lead to mucolipidosis type IV, a severe lysosomal storage disorder. Here we report two electron cryo-microscopy structures of full-length human TRPML1: a 3.72-Å apo structure at pH 7.0 in the closed state, and a 3.49-Å agonist-bound structure at pH 6.0 in an open state. Several aromatic and hydrophobic residues in pore helix 1, helices S5 and S6, and helix S6 of a neighbouring subunit, form a hydrophobic cavity to house the agonist, suggesting a distinct agonist-binding site from that found in TRPV1, a TRP channel from a different subfamily. The opening of TRPML1 is associated with distinct dilations of its lower gate together with a slight structural movement of pore helix 1. Our work reveals the regulatory mechanism of TRPML channels, facilitates better understanding of TRP channel activation, and provides insights into the molecular basis of mucolipidosis type IV pathogenesis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Zeevi, D. A., Frumkin, A. & Bach, G. TRPML and lysosomal function. Biochim. Biophys. Acta 1772, 851–858 (2007)
Xu, H. & Ren, D. Lysosomal physiology. Annu. Rev. Physiol. 77, 57–80 (2015)
Venkatachalam, K., Wong, C. O. & Zhu, M. X. The role of TRPMLs in endolysosomal trafficking and function. Cell Calcium 58, 48–56 (2015)
Samie, M. et al. A TRP channel in the lysosome regulates large particle phagocytosis via focal exocytosis. Dev. Cell 26, 511–524 (2013)
Miedel, M. T. et al. Membrane traffic and turnover in TRP-ML1-deficient cells: a revised model for mucolipidosis type IV pathogenesis. J. Exp. Med. 205, 1477–1490 (2008)
Li, X. et al. A molecular mechanism to regulate lysosome motility for lysosome positioning and tubulation. Nat. Cell Biol. 18, 404–417 (2016)
Venkatachalam, K. et al. Motor deficit in a Drosophila model of mucolipidosis type IV due to defective clearance of apoptotic cells. Cell 135, 838–851 (2008)
Vergarajauregui, S., Connelly, P. S., Daniels, M. P. & Puertollano, R. Autophagic dysfunction in mucolipidosis type IV patients. Hum. Mol. Genet. 17, 2723–2737 (2008)
Zhang, X. et al. MCOLN1 is a ROS sensor in lysosomes that regulates autophagy. Nat. Commun. 7, 12109 (2016)
Dong, X. P., Wang, X. & Xu, H. TRP channels of intracellular membranes. J. Neurochem. 113, 313–328 (2010)
Soyombo, A. A. et al. TRP-ML1 regulates lysosomal pH and acidic lysosomal lipid hydrolytic activity. J. Biol. Chem. 281, 7294–7301 (2006)
Shen, D. et al. Lipid storage disorders block lysosomal trafficking by inhibiting a TRP channel and lysosomal calcium release. Nat. Commun. 3, 731 (2012)
Chen, C. C. et al. A small molecule restores function to TRPML1 mutant isoforms responsible for mucolipidosis type IV. Nat. Commun. 5, 4681 (2014)
Zhang, X., Li, X. & Xu, H. Phosphoinositide isoforms determine compartment-specific ion channel activity. Proc. Natl Acad. Sci. USA 109, 11384–11389 (2012)
Dong, X. P. et al. PI(3,5)P2 controls membrane trafficking by direct activation of mucolipin Ca2+ release channels in the endolysosome. Nat. Commun. 1, 38 (2010)
Wang, W., Zhang, X., Gao, Q. & Xu, H. TRPML1: an ion channel in the lysosome. Handb. Exp. Pharmacol. 222, 631–645 (2014)
Sun, M. et al. Mucolipidosis type IV is caused by mutations in a gene encoding a novel transient receptor potential channel. Hum. Mol. Genet. 9, 2471–2478 (2000)
Bargal, R. et al. Identification of the gene causing mucolipidosis type IV. Nat. Genet. 26, 118–123 (2000)
Bassi, M. T. et al. Cloning of the gene encoding a novel integral membrane protein, mucolipidin—and identification of the two major founder mutations causing mucolipidosis type IV. Am. J. Hum. Genet. 67, 1110–1120 (2000)
Weitz, R. & Kohn, G. Clinical spectrum of mucolipidosis type IV. Pediatrics 81, 602–603 (1988)
Bach, G. Mucolipidosis type IV. Mol. Genet. Metab. 73, 197–203 (2001)
Venkatachalam, K. & Montell, C. TRP channels. Annu. Rev. Biochem. 76, 387–417 (2007)
García-Añoveros, J. & Wiwatpanit, T. TRPML2 and mucolipin evolution. Handb. Exp. Pharmacol. 222, 647–658 (2014)
Grimm, C., Barthmes, M. & Wahl-Schott, C. Trpml3. Handb. Exp. Pharmacol. 222, 659–674 (2014)
Di Palma, F. et al. Mutations in Mcoln3 associated with deafness and pigmentation defects in varitint-waddler (Va) mice. Proc. Natl Acad. Sci. USA 99, 14994–14999 (2002)
Cao, E., Liao, M., Cheng, Y. & Julius, D. TRPV1 structures in distinct conformations reveal activation mechanisms. Nature 504, 113–118 (2013)
Liao, M., Cao, E., Julius, D. & Cheng, Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature 504, 107–112 (2013)
Paulsen, C. E., Armache, J. P., Gao, Y., Cheng, Y. & Julius, D. Structure of the TRPA1 ion channel suggests regulatory mechanisms. Nature 520, 511–517 (2015)
Gao, Y., Cao, E., Julius, D. & Cheng, Y. TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature 534, 347–351 (2016)
Zubcevic, L. et al. Cryo-electron microscopy structure of the TRPV2 ion channel. Nat. Struct. Mol. Biol. 23, 180–186 (2016)
Huynh, K. W. et al. Structure of the full-length TRPV2 channel by cryo-EM. Nat. Commun. 7, 11130 (2016)
Shen, P. S. et al. The structure of the polycystic kidney disease channel PKD2 in lipid nanodiscs. Cell 167, 763–773.e11, (2016)
Wilkes, M. et al. Molecular insights into lipid-assisted Ca2+ regulation of the TRP channel Polycystin-2. Nat. Struct. Mol. Biol. 24, 123–130 (2017)
Grieben, M. et al. Structure of the polycystic kidney disease TRP channel Polycystin-2 (PC2). Nat. Struct. Mol. Biol. 24, 114–122 (2017)
Li, M. et al. Structural basis of dual Ca2+/pH regulation of the endolysosomal TRPML1 channel. Nat. Struct. Mol. Biol. 24, 205–213 (2017)
Goehring, A. et al. Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat. Protocols 9, 2574–2585 (2014)
Tang, L. et al. Structural basis for inhibition of a voltage-gated Ca2+ channel by Ca2+ antagonist drugs. Nature 537, 117–121 (2016)
Vergarajauregui, S. & Puertollano, R. Two di-leucine motifs regulate trafficking of mucolipin-1 to lysosomes. Traffic 7, 337–353 (2006)
Dong, X. P. et al. Activating mutations of the TRPML1 channel revealed by proline-scanning mutagenesis. J. Biol. Chem. 284, 32040–32052 (2009)
Li, X., Saha, P., Li, J., Blobel, G. & Pfeffer, S. R. Clues to the mechanism of cholesterol transfer from the structure of NPC1 middle lumenal domain bound to NPC2. Proc. Natl Acad. Sci. USA 113, 10079–10084 (2016)
Pipalia, N. H. et al. Histone deacetylase inhibitors correct the cholesterol storage defect in most Niemann-Pick C1 mutant cells. J. Lipid Res. 58, 695–708 (2017)
Chen, Q. et al. Structure of mammalian endolysosomal TRPML1 channel in nanodiscs. Nature http://doi.org/10.1038/nature24035 (2017)
Fine, M. et al. Massive endocytosis driven by lipidic forces originating in the outer plasmalemmal monolayer: a new approach to membrane recycling and lipid domains. J. Gen. Physiol. 137, 137–154 (2011)
Wang, T. M. & Hilgemann, D. W. Ca-dependent nonsecretory vesicle fusion in a secretory cell. J. Gen. Physiol. 132, 51–65 (2008)
Grant, T. & Grigorieff, N. Measuring the optimal exposure for single particle cryo-EM using a 2.6 Å reconstruction of rotavirus VP6. eLife 4, e06980 (2015)
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015)
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012)
Rubinstein, J. L. & Brubaker, M. A. Alignment of cryo-EM movies of individual particles by optimization of image translations. J. Struct. Biol. 192, 188–195 (2015)
Grigorieff, N. Frealign: an exploratory tool for single-particle cryo-EM. Methods Enzymol. 579, 191–226 (2016)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997)
Brown, A. et al. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr. D 71, 136–153 (2015)
Ten Eyck, L. F. Efficient structure-factor calculation for large molecules by the fast Fourier transform. Acta Crystallogr. A 33, 486–492 (1977)
Wang, Z. et al. An atomic model of brome mosaic virus using direct electron detection and real-space optimization. Nat. Commun. 5, 4808 (2014)
Heymann, J. B. & Belnap, D. M. Bsoft: image processing and molecular modeling for electron microscopy. J. Struct. Biol. 157, 3–18 (2007)
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010)
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004)
Smart, O. S., Neduvelil, J. G., Wang, X., Wallace, B. A. & Sansom, M. S. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14, 354–360, 376 (1996)
Acknowledgements
We thank M. Ebrahim and J. Sotiris at the Rockefeller Cryo-EM Resource Center for assistance in data collection, Z. Zhang for help in EM data processing, D. Hilgemann, E. Coutavas and E. Debler for help in manuscript preparation, and J. Fernandez and H. Molina for mass spectrometry analyses. This work was supported by funds from the Howard Hughes Medical Institute (to G.B.) and the Endowed Scholars Program in Medical Science of University of Texas Southwestern Medical Center (to X.L.). M.F. was supported by the National Institutes of Health (T32DK007257 and HL119843). X.L. was the recipient of a Gordon and Betty Moore Foundation Fellow of the Life Sciences Research Foundation.
Author information
Authors and Affiliations
Contributions
P.S. and X.L. purified proteins and carried out cryo-EM work. M.F. carried out functional characterization by electrophysiology. All authors analysed data and contributed to manuscript preparation. X.L. conceived the project and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks S. Hansen, C. Ulens and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 Sequence alignment of human TRPML1, TRPML2 and TRPML3.
Residues discussed in the paper are annotated using symbols at their positions.
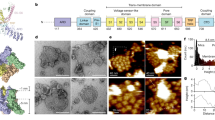
Extended Data Figure 2 Biochemical properties and cryo-EM studies of human TRPML1.
a, Size-exclusion chromatogram and SDS–PAGE gel of the purified TRPML1. A degradation fragment is presented on the gel and is indicated by an asterisk. b, Density maps of structures coloured by local resolution estimation using blocres56.
Extended Data Figure 3 Data and model quality assessment.
a, Left to right, a representative electron micrograph at defocus −2.0 μm; 2D classification; and FSC curves of the apo structure. The left curve shows a plot of the FSC as a function of resolution using Frealign output, the right curve shows the FSC calculated between the refined structure and the half map used for refinement, the other half map, and the full map. b, Left to right, a representative micrograph at defocus −2.0 μm; 2D classification; and FSC curves of ML-SA1-bound TRPML1 structure. The two FSC curves represent the same as for panel a.
Extended Data Figure 4 Electron microscopy density of different portions of the structures.
a, The apo TRPML1 structure. b, The ML-SA1-bound TRPML1 structure.
Extended Data Figure 5 Comparisons of TRPML1 and PKD2.
a, Superimposition of overall structures of TRPML1 and PKD2 (PDB code: 5T4D). b, Superimposition of one subunit of TRPML1 and PKD2. The extended structural elements of pre-S1 (α1 and α2), and α4 and S2 of TRPML1, are highlighted in purple and pink, respectively.
Extended Data Figure 6 Whole-cell currents of HEK wild-type cells and TRPML1(L/A) with Y436A, F465A or Y499A mutations.
a, Whole-cell currents of HEK293T cells transfected with empty vector at pH 4.6 or pH 7.2 with or without 10 μM ML-SA1 and 50 μΜ PtdIns(3,5)P2. b, Whole-cell currents of HEK293T cells transfected with surface-expressing mutant eGFP–TRPML1(L/A) containing Y436A, F465A or Y499A under the same conditions as in panel a. c, Confocal images of patched cells. Scale bars, 10 μm.
Extended Data Figure 7 The distribution of mutations that cause mucolipidosis type IV in the TRPML1 structure.
The mutations are shown as grey balls.
Supplementary information
Rights and permissions
About this article
Cite this article
Schmiege, P., Fine, M., Blobel, G. et al. Human TRPML1 channel structures in open and closed conformations. Nature 550, 366–370 (2017). https://doi.org/10.1038/nature24036
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature24036
This article is cited by
-
TRP (transient receptor potential) ion channel family: structures, biological functions and therapeutic interventions for diseases
Signal Transduction and Targeted Therapy (2023)
-
Cryo-EM structure of the human Kv3.1 channel reveals gating control by the cytoplasmic T1 domain
Nature Communications (2022)
-
Release and uptake mechanisms of vesicular Ca2+ stores
Protein & Cell (2019)
-
Structural basis for PtdInsP2-mediated human TRPML1 regulation
Nature Communications (2018)
-
Cryo-EM in drug discovery: achievements, limitations and prospects
Nature Reviews Drug Discovery (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.