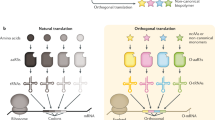
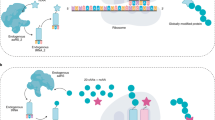
Abstract
Nature uses a limited, conservative set of amino acids to synthesize proteins. The ability to genetically encode an expanded set of building blocks with new chemical and physical properties is transforming the study, manipulation and evolution of proteins, and is enabling diverse applications, including approaches to probe, image and control protein function, and to precisely engineer therapeutics. Underpinning this transformation are strategies to engineer and rewire translation. Emerging strategies aim to reprogram the genetic code so that noncanonical biopolymers can be synthesized and evolved, and to test the limits of our ability to engineer the translational machinery and systematically recode genomes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Ambrogelly, A., Palioura, S. & Söll, D. Natural expansion of the genetic code. Nat. Chem. Biol. 3, 29–35 (2007)
Dougherty, D. A. & Van Arnam, E. B. In vivo incorporation of non-canonical amino acids by using the chemical aminoacylation strategy: a broadly applicable mechanistic tool. ChemBioChem 15, 1710–1720 (2014)
Cornish, V. W., Mendel, D. & Schultz, P. G. Probing protein structure and function with an expanded genetic code. Angew. Chem. Int. Edn Engl. 34, 621–633 (1995)
Bondalapati, S., Jbara, M. & Brik, A. Expanding the chemical toolbox for the synthesis of large and uniquely modified proteins. Nat. Chem. 8, 407–418 (2016)
Chin, J. W. Expanding and reprogramming the genetic code of cells and animals. Annu. Rev. Biochem. 83, 379–408 (2014)
Liu, C. C. & Schultz, P. G. Adding new chemistries to the genetic code. Annu. Rev. Biochem. 79, 413–444 (2010)
Zhang, M. S . et al. Biosynthesis and genetic encoding of phosphothreonine through parallel selection and deep sequencing. Nat. Methods 14, 729–736 (2017). Describes a scalable approach to the discovery of orthgonal synthetases through parallel positive selection and sequencing and a strategy to biosynthesize and encode a key post-translational modification.
Anderson, J. C. & Schultz, P. G. Adaptation of an orthogonal archaeal leucyl-tRNA and synthetase pair for four-base, amber, and opal suppression. Biochemistry 42, 9598–9608 (2003)
Neumann, H., Peak-Chew, S. Y. & Chin, J. W. Genetically encoding N(ε)-acetyllysine in recombinant proteins. Nat. Chem. Biol. 4, 232–234 (2008)
Ellefson, J. W. et al. Directed evolution of genetic parts and circuits by compartmentalized partnered replication. Nat. Biotechnol. 32, 97–101 (2014)
Wang, L., Brock, A., Herberich, B. & Schultz, P. G. Expanding the genetic code of Escherichia coli. Science 292, 498–500 (2001)
Young, D. D. et al. An evolved aminoacyl-tRNA synthetase with atypical polysubstrate specificity. Biochemistry 50, 1894–1900 (2011)
Cooley, R. B., Karplus, P. A. & Mehl, R. A. Gleaning unexpected fruits from hard-won synthetases: probing principles of permissivity in non-canonical amino acid-tRNA synthetases. ChemBioChem 15, 1810–1819 (2014)
Dumas, A., Lercher, L., Spicer, C. D. & Davis, B. G. Designing logical codon reassignment — Expanding the chemistry in biology. Chem. Sci. 6, 50–69 (2015)
Santoro, S. W., Anderson, J. C., Lakshman, V. & Schultz, P. G. An archaebacteria-derived glutamyl-tRNA synthetase and tRNA pair for unnatural amino acid mutagenesis of proteins in Escherichia coli. Nucleic Acids Res. 31, 6700–6709 (2003)
Tes¸ileanu, T., Colwell, L. J. & Leibler, S. Protein sectors: statistical coupling analysis versus conservation. PLOS Comput. Biol. 11, e1004091 (2015)
Iraha, F. et al. Functional replacement of the endogenous tyrosyl-tRNA synthetase-tRNATyr pair by the archaeal tyrosine pair in Escherichia coli for genetic code expansion. Nucleic Acids Res. 38, 3682–3691 (2010)
Hughes, R. A. & Ellington, A. D. Rational design of an orthogonal tryptophanyl nonsense suppressor tRNA. Nucleic Acids Res. 38, 6813–6830 (2010)
Italia, J. S . et al. An orthogonalized platform for genetic code expansion in both bacteria and eukaryotes. Nat. Chem. Biol. 13, 446–450 (2017). Evolution of the E. coli TrpRS–tRNA pair in E. coli and transfer of this pair to mammalian cells for genetic code expansion.
Ernst, R. J. et al. Genetic code expansion in the mouse brain. Nat. Chem. Biol. 12, 776–778 (2016)
Han, S. et al. Expanding the genetic code of Mus musculus. Nat. Commun. 8, 14568 (2017)
Elliott, T. S. et al. Proteome labeling and protein identification in specific tissues and at specific developmental stages in an animal. Nat. Biotechnol. 32, 465–472 (2014)
Elliott, T. S., Bianco, A., Townsley, F. M., Fried, S. D. & Chin, J. W. Tagging and enriching proteins enables cell-specific proteomics. Cell Chem. Biol. 23, 805–815 (2016)
Stone, S. E., Glenn, W. S., Hamblin, G. D. & Tirrell, D. A. Cell-selective proteomics for biological discovery. Curr. Opin. Chem. Biol. 36, 50–57 (2017)
Schmied, W. H., Elsässer, S. J., Uttamapinant, C. & Chin, J. W. Efficient multisite unnatural amino acid incorporation in mammalian cells via optimized pyrrolysyl tRNA synthetase/tRNA expression and engineered eRF1. J. Am. Chem. Soc. 136, 15577–15583 (2014)
Amiram, M. et al. Evolution of translation machinery in recoded bacteria enables multi-site incorporation of nonstandard amino acids. Nat. Biotechnol. 33, 1272–1279 (2015)
Zheng, Y., Lewis, T. L., Jr, Igo, P., Polleux, F. & Chatterjee, A. Virus-enabled optimization and delivery of the genetic machinery for efficient unnatural amino acid mutagenesis in mammalian cells and tissues. ACS Synth. Biol. 6, 13–18 (2017)
Chatterjee, A., Sun, S. B., Furman, J. L., Xiao, H. & Schultz, P. G. A versatile platform for single- and multiple-unnatural amino acid mutagenesis in Escherichia coli. Biochemistry 52, 1828–1837 (2013)
Wang, K., Neumann, H., Peak-Chew, S. Y. & Chin, J. W. Evolved orthogonal ribosomes enhance the efficiency of synthetic genetic code expansion. Nat. Biotechnol. 25, 770–777 (2007)
Neumann, H., Wang, K., Davis, L., Garcia-Alai, M. & Chin, J. W. Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature 464, 441–444 (2010)
Wang, K. et al. Optimized orthogonal translation of unnatural amino acids enables spontaneous protein double-labelling and FRET. Nat. Chem. 6, 393–403 (2014)
Park, H. S. et al. Expanding the genetic code of Escherichia coli with phosphoserine. Science 333, 1151–1154 (2011)
Fan, C., Ip, K. & Söll, D. Expanding the genetic code of Escherichia coli with phosphotyrosine. FEBS Lett. 590, 3040–3047 (2016)
Mukai, T. et al. Codon reassignment in the Escherichia coli genetic code. Nucleic Acids Res. 38, 8188–8195 (2010)
Johnson, D. B. et al. RF1 knockout allows ribosomal incorporation of unnatural amino acids at multiple sites. Nat. Chem. Biol. 7, 779–786 (2011)
Lajoie, M. J. et al. Genomically recoded organisms expand biological functions. Science 342, 357–360 (2013)
Wu, I. L. et al. Multiple site-selective insertions of noncanonical amino acids into sequence-repetitive polypeptides. ChemBioChem 14, 968–978 (2013)
Chatterjee, A., Lajoie, M. J., Xiao, H., Church, G. M. & Schultz, P. G. A bacterial strain with a unique quadruplet codon specifying non-native amino acids. ChemBioChem 15, 1782–1786 (2014)
Mukai, T. et al. Highly reproductive Escherichia coli cells with no specific assignment to the UAG codon. Sci. Rep. 5, 9699 (2015)
Zheng, Y. et al. Performance of optimized noncanonical amino acid mutagenesis systems in the absence of release factor 1. Mol. Biosyst. 12, 1746–1749 (2016)
Rogerson, D. T. et al. Efficient genetic encoding of phosphoserine and its nonhydrolyzable analog. Nat. Chem. Biol. 11, 496–503 (2015)
Richardson, S. M. et al. Design of a synthetic yeast genome. Science 355, 1040–1044 (2017)
Davis, L. & Chin, J. W. Designer proteins: applications of genetic code expansion in cell biology. Nat. Rev. Mol. Cell Biol. 13, 168–182 (2012)
Minnihan, E. C., Seyedsayamdost, M. R., Uhlin, U. & Stubbe, J. Kinetics of radical intermediate formation and deoxynucleotide production in 3-aminotyrosine-substituted Escherichia coli ribonucleotide reductases. J. Am. Chem. Soc. 133, 9430–9440 (2011)
Yang, Y. et al. Genetically encoded protein photocrosslinker with a transferable mass spectrometry-identifiable label. Nat. Commun. 7, 12299 (2016)
Coin, I. et al. Genetically encoded chemical probes in cells reveal the binding path of urocortin-I to CRF class B GPCR. Cell 155, 1258–1269 (2013)
Lang, K. & Chin, J. W. Cellular incorporation of unnatural amino acids and bioorthogonal labeling of proteins. Chem. Rev. 114, 4764–4806 (2014)
Chatterjee, A., Guo, J., Lee, H. S. & Schultz, P. G. A genetically encoded fluorescent probe in mammalian cells. J. Am. Chem. Soc. 135, 12540–12543 (2013)
Luo, J. et al. Genetically encoded optochemical probes for simultaneous fluorescence reporting and light activation of protein function with two-photon excitation. J. Am. Chem. Soc. 136, 15551–15558 (2014)
Lang, K. & Chin, J. W. Bioorthogonal reactions for labeling proteins. ACS Chem. Biol. 9, 16–20 (2014)
Lukinavicˇius, G. et al. A near-infrared fluorophore for live-cell super-resolution microscopy of cellular proteins. Nat. Chem. 5, 132–139 (2013)
Uttamapinant, C. et al. Genetic code expansion enables live-cell and super-resolution imaging of site-specifically labeled cellular proteins. J. Am. Chem. Soc. 137, 4602–4605 (2015)
Kipper, K. et al. Application of noncanonical amino acids for protein labeling in a genomically recoded Escherichia coli. ACS Synth. Biol. 6, 233–255 (2017)
Peng, T. & Hang, H. C. Site-specific bioorthogonal labeling for fluorescence imaging of intracellular proteins in living cells. J. Am. Chem. Soc. 138, 14423–14433 (2016)
Baumdick, M. et al. EGF-dependent re-routing of vesicular recycling switches spontaneous phosphorylation suppression to EGFR signaling. eLife 4, e12223 (2015)
Sakin, V. et al. A versatile tool for live-cell imaging and super-resolution nanoscopy studies of HIV-1 Env distribution and mobility. Cell. Chem. Biol. 24, 635–645 (2017)
Xue, L., Prifti, E. & Johnsson, K. A general strategy for the semisynthesis of ratiometric fluorescent sensor proteins with increased dynamic range. J. Am. Chem. Soc. 138, 5258–5261 (2016)
Sachdeva, A., Wang, K., Elliott, T. & Chin, J. W. Concerted, rapid, quantitative, and site-specific dual labeling of proteins. J. Am. Chem. Soc. 136, 7785–7788 (2014)
Xiao, H. et al. Genetic incorporation of multiple unnatural amino acids into proteins in mammalian cells. Angew. Chem. Int. Edn Engl. 52, 14080–14083 (2013)
Li, J. et al. Palladium-triggered deprotection chemistry for protein activation in living cells. Nat. Chem. 6, 352–361 (2014)
Li, J., Jia, S. & Chen, P. R. Diels–Alder reaction-triggered bioorthogonal protein decaging in living cells. Nat. Chem. Biol. 10, 1003–1005 (2014)
Zhang, G. et al. Bioorthogonal chemical activation of kinases in living systems. ACS Cent. Sci. 2, 325–331 (2016)
Wang, J. et al. Palladium-triggered chemical rescue of intracellular proteins via genetically encoded allene-caged tyrosine. J. Am. Chem. Soc. 138, 15118–15121 (2016)
Baker, A. S. & Deiters, A. Optical control of protein function through unnatural amino acid mutagenesis and other optogenetic approaches. ACS Chem. Biol. 9, 1398–1407 (2014)
Hemphill, J., Borchardt, E. K., Brown, K., Asokan, A. & Deiters, A. Optical control of CRISPR/Cas9 gene editing. J. Am. Chem. Soc. 137, 5642–5645 (2015)
Nguyen, D. P. et al. Genetic encoding of photocaged cysteine allows photoactivation of TEV protease in live mammalian cells. J. Am. Chem. Soc. 136, 2240–2243 (2014)
Walker, O. S. et al. Photoactivation of mutant isocitrate dehydrogenase 2 reveals rapid cancer-associated metabolic and epigenetic changes. J. Am. Chem. Soc. 138, 718–721 (2016)
Bose, M., Groff, D., Xie, J., Brustad, E. & Schultz, P. G. The incorporation of a photoisomerizable amino acid into proteins in E. coli. J. Am. Chem. Soc. 128, 388–389 (2006)
Hoppmann, C., Maslennikov, I., Choe, S. & Wang, L. In situ formation of an azo bridge on proteins controllable by visible light. J. Am. Chem. Soc. 137, 11218–11221 (2015)
Tsai, Y.-H., Essig, S., James, J. R., Lang, K. & Chin, J. W. Selective, rapid and optically switchable regulation of protein function in live mammalian cells. Nat. Chem. 7, 554–561 (2015)
Elsässer, S. J., Ernst, R. J., Walker, O. S. & Chin, J. W. Genetic code expansion in stable cell lines enables encoded chromatin modification. Nat. Methods 13, 158–164 (2016)
Si, L . et al. Generation of influenza A viruses as live but replication-incompetent virus vaccines. Science 354, 1170–1173 (2016). Describes the creation of an amber suppression-dependent influenza A virus and the use of the resulting attenuated virus for immunization.
Wang, Z. A. et al. A genetically encoded allysine for the synthesis of proteins with site-specific lysine dimethylation. Angew. Chem. Int. Edn Engl. 56, 212–216 (2017)
Wang, Z. A. et al. A versatile approach for site-specific lysine acylation in proteins. Angew. Chem. Int. Edn Engl. 56, 1643–1647 (2017)
Hoppmann, C. et al. Site-specific incorporation of phosphotyrosine using an expanded genetic code. Nat. Chem. Biol. 13, 842–844 (2017)
Luo, X. et al. Genetically encoding phosphotyrosine and its nonhydrolyzable analog in bacteria. Nat. Chem. Biol. 13, 845–849 (2017)
Wright, T. H. et al. Posttranslational mutagenesis: a chemical strategy for exploring protein side-chain diversity. Science 354, aag1465 (2016)
Yang, A. et al. A chemical biology route to site-specific authentic protein modifications. Science 354, 623–626 (2016)
Xiao, H. & Schultz, P. G. At the interface of chemical and biological synthesis: an expanded genetic code. Cold Spring Harb. Perspect. Biol. 8, a023945 (2016)
Tian, F. et al. A general approach to site-specific antibody drug conjugates. Proc. Natl Acad. Sci. USA 111, 1766–1771 (2014)
VanBrunt, M. P. et al. Genetically encoded azide containing amino acid in mammalian cells enables site-specific antibody-drug conjugates using click cycloaddition chemistry. Bioconjug. Chem. 26, 2249–2260 (2015)
Kularatne, S. A. et al. Recruiting cytotoxic T cells to folate-receptor-positive cancer cells. Angew. Chem. Int. Edn Engl. 52, 12101–12104 (2013)
Ma, J. S. et al. Versatile strategy for controlling the specificity and activity of engineered T cells. Proc. Natl Acad. Sci. USA 113, E450–E458 (2016)
Wang, N. et al. Construction of a live-attenuated HIV-1 vaccine through genetic code expansion. Angew. Chem. Int. Edn Engl. 53, 4867–4871 (2014)
Lin, S. et al. Site-specific engineering of chemical functionalities on the surface of live hepatitis D virus. Angew. Chem. Int. Edn Engl. 52, 13970–13974 (2013)
Loughran, G. et al. Evidence of efficient stop codon readthrough in four mammalian genes. Nucleic Acids Res. 42, 8928–8938 (2014)
Xuan, W. & Schultz, P. G. A strategy for creating organisms dependent on noncanonical amino acids. Angew. Chem. Int. Edn Engl. 56, 9170–9173 (2017)
Mandell, D. J. et al. Biocontainment of genetically modified organisms by synthetic protein design. Nature 518, 55–60 (2015). Makes the function of essential genes dependent on both amber suppression and the identity of a noncanonical amino acid incorporated by amber suppression.
Mills, J. H. et al. Computational design of an unnatural amino acid dependent metalloprotein with atomic level accuracy. J. Am. Chem. Soc. 135, 13393–13399 (2013)
Mills, J. H. et al. Computational design of a homotrimeric metalloprotein with a trisbipyridyl core. Proc. Natl Acad. Sci. USA 113, 15012–15017 (2016). Design of metalloproteins with an expanded genetic code.
Pearson, A. D. et al. Trapping a transition state in a computationally designed protein bottle. Science 347, 863–867 (2015)
Xiao, H. et al. Exploring the potential impact of an expanded genetic code on protein function. Proc. Natl Acad. Sci. USA 112, 6961–6966 (2015)
Tack, D. S. et al. Addicting diverse bacteria to a noncanonical amino acid. Nat. Chem. Biol. 12, 138–140 (2016)
Hammerling, M. J. et al. Bacteriophages use an expanded genetic code on evolutionary paths to higher fitness. Nat. Chem. Biol. 10, 178–180 (2014)
Lopez, G. & Anderson, J. C. Synthetic auxotrophs with ligand-dependent essential genes for a BL21(DE3) biosafety strain. ACS Synth. Biol. 4, 1279–1286 (2015)
Forster, A. C. et al. Programming peptidomimetic syntheses by translating genetic codes designed de novo. Proc. Natl Acad. Sci. USA 100, 6353–6357 (2003)
Iwane, Y. et al. Expanding the amino acid repertoire of ribosomal polypeptide synthesis via the artificial division of codon boxes. Nat. Chem. 8, 317–325 (2016)
Anderson, J. C. et al. An expanded genetic code with a functional quadruplet codon. Proc. Natl Acad. Sci. USA 101, 7566–7571 (2004)
Malyshev, D. A. et al. A semi-synthetic organism with an expanded genetic alphabet. Nature 509, 385–388 (2014)
Zhang, Y . et al. A semisynthetic organism engineered for the stable expansion of the genetic alphabet. Proc. Natl Acad. Sci. USA 114, 1317–1322 (2017). The maintenance of an orthogonal base pair in E. coli.
Ngo, J. T. & Tirrell, D. A. Noncanonical amino acids in the interrogation of cellular protein synthesis. Acc. Chem. Res. 44, 677–685 (2011)
Zeng, Y., Wang, W. & Liu, W. R. Towards reassigning the rare AGG codon in Escherichia coli. ChemBioChem 15, 1750–1754 (2014)
Ostrov, N . et al. Design, synthesis, and testing toward a 57-codon genome. Science 353, 819–822 (2016). Replaces 50-kb sections of the E. coli genome in ten independent strains using variable synonymous codon replacements.
Wang, K. et al. Defining synonymous codon compression schemes by genome recoding. Nature 539, 59–64 (2016). Describes a powerful strategy for genome replacement in E. coli and its application to deciphering the best synonymous substitutions for codons targeted for removal from the genome.
Napolitano, M. G. et al. Emergent rules for codon choice elucidated by editing rare arginine codons in Escherichia coli. Proc. Natl Acad. Sci. USA 113, E5588–E5597 (2016)
Lau, Y. H. et al. Large-scale recoding of a bacterial genome by iterative recombineering of synthetic DNA. Nucleic Acids Res. 45, 6971–6980 (2017)
Neumann, H., Slusarczyk, A. L. & Chin, J. W. De novo generation of mutually orthogonal aminoacyl-tRNA synthetase/tRNA pairs. J. Am. Chem. Soc. 132, 2142–2144 (2010)
Chatterjee, A., Xiao, H. & Schultz, P. G. Evolution of multiple, mutually orthogonal prolyl-tRNA synthetase/tRNA pairs for unnatural amino acid mutagenesis in Escherichia coli. Proc. Natl Acad. Sci. USA 109, 14841–14846 (2012)
Guo, J., Wang, J., Anderson, J. C. & Schultz, P. G. Addition of an alpha-hydroxy acid to the genetic code of bacteria. Angew. Chem. Int. Edn Engl. 47, 722–725 (2008)
Kobayashi, T., Yanagisawa, T., Sakamoto, K. & Yokoyama, S. Recognition of non-alpha-amino substrates by pyrrolysyl-tRNA synthetase. J. Mol. Biol. 385, 1352–1360 (2009)
Tan, Z., Forster, A. C., Blacklow, S. C. & Cornish, V. W. Amino acid backbone specificity of the Escherichia coli translation machinery. J. Am. Chem. Soc. 126, 12752–12753 (2004)
Fujino, T., Goto, Y., Suga, H. & Murakami, H. Reevaluation of the d-amino acid compatibility with the elongation event in translation. J. Am. Chem. Soc. 135, 1830–1837 (2013)
Maini, R. et al. Protein synthesis with ribosomes selected for the incorporation of β-amino acids. Biochemistry 54, 3694–3706 (2015)
Maini, R. et al. Ribosome-mediated incorporation of dipeptides and dipeptide analogues into proteins in vitro. J. Am. Chem. Soc. 137, 11206–11209 (2015)
Dedkova, L. M., Fahmi, N. E., Golovine, S. Y. & Hecht, S. M. Construction of modified ribosomes for incorporation of d-amino acids into proteins. Biochemistry 45, 15541–15551 (2006)
Melo Czekster, C., Robertson, W. E., Walker, A. S., Söll, D. & Schepartz, A. In vivo biosynthesis of a β-amino acid-containing protein. J. Am. Chem. Soc. 138, 5194–5197 (2016)
Englander, M. T. et al. The ribosome can discriminate the chirality of amino acids within its peptidyl-transferase center. Proc. Natl Acad. Sci. USA 112, 6038–6043 (2015)
Terasaka, N., Hayashi, G., Katoh, T. & Suga, H. An orthogonal ribosome-tRNA pair via engineering of the peptidyl transferase center. Nat. Chem. Biol. 10, 555–557 (2014)
Fried, S. D., Schmied, W. H., Uttamapinant, C. & Chin, J. W. Ribosome subunit stapling for orthogonal translation in E. coli. Angew. Chem. Int. Edn Engl. 54, 12791–12794 (2015)
Orelle, C. et al. Protein synthesis by ribosomes with tethered subunits. Nature 524, 119–124 (2015)
Acknowledgements
I thank A. Chatterjee, K. Wang, and P. G. Schultz for suggestions and edits to earlier versions of the manuscript and V. Beranek, and K. Wang for assistance with figures. Work in my laboratory is supported by the Medical Research Council, UK (MC_U105181009 and MC_UP_A024_1008) and an ERC Advanced Grant (SGCR).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The author declares no competing financial interests.
Additional information
Reviewer Information Nature thanks A. C. Forster, A. Schepartz and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Chin, J. Expanding and reprogramming the genetic code. Nature 550, 53–60 (2017). https://doi.org/10.1038/nature24031
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature24031
This article is cited by
-
Blocking and rescuing tryptophan interactions
Nature Chemistry (2024)
-
Adeno-associated virus as a delivery vector for gene therapy of human diseases
Signal Transduction and Targeted Therapy (2024)
-
Irreversible light-activated SpyLigation mediates split-protein assembly in 4D
Nature Protocols (2024)
-
Electrochemical labelling of hydroxyindoles with chemoselectivity for site-specific protein bioconjugation
Nature Chemistry (2024)
-
Adding α,α-disubstituted and β-linked monomers to the genetic code of an organism
Nature (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.