Abstract
Epithelial organs undergo steady-state turnover throughout adult life, with old cells being continually replaced by the progeny of stem cell divisions1. To avoid hyperplasia or atrophy, organ turnover demands strict equilibration of cell production and loss2,3,4. However, the mechanistic basis of this equilibrium is unknown. Here we show that robustly precise turnover of the adult Drosophila intestine5 arises through a coupling mechanism in which enterocyte apoptosis breaks feedback inhibition of stem cell division. Healthy enterocytes inhibit stem cell division through E-cadherin, which prevents secretion of mitogenic epidermal growth factors (EGFs) by repressing transcription of the EGF maturation factor rhomboid. Individual apoptotic enterocytes promote divisions by loss of E-cadherin, which releases cadherin-associated β-catenin (Armadillo in Drosophila) and p120-catenin to induce rhomboid. Induction of rhomboid in the dying enterocyte triggers activation of the EGF receptor (Egfr) in stem cells within a discrete radius. When we blocked apoptosis, E-cadherin-controlled feedback suppressed divisions, and the organ retained the same number of cells. When we disrupted feedback, apoptosis and divisions were uncoupled, and the organ developed either hyperplasia or atrophy. Together, our results show that robust cellular balance hinges on the obligate coupling of divisions to apoptosis, which limits the proliferative potential of a stem cell to the precise time and place at which a replacement cell is needed. In this way, localized cell–cell communication gives rise to tissue-level homeostatic equilibrium and constant organ size.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Leblond, C. P. & Stevens, C. E. The constant renewal of the intestinal epithelium in the albino rat. Anat. Rec. 100, 357–377 (1948)
Pellettieri, J. & Sánchez Alvarado, A. Cell turnover and adult tissue homeostasis: from humans to planarians. Annu. Rev. Genet. 41, 83–105 (2007)
O’Brien, L. E., Soliman, S. S., Li, X. & Bilder, D. Altered modes of stem cell division drive adaptive intestinal growth. Cell 147, 603–614 (2011)
O’Brien, L. E. & Bilder, D. Beyond the niche: tissue-level coordination of stem cell dynamics. Annu. Rev. Cell Dev. Biol. 29, 107–136 (2013)
Apidianakis, Y., Tamamouna, V., Teloni, S. & Pitsouli, C. in Advances in Insect Physiology Vol. 52 (ed. Ligoxygakis, P. ) 139–178 (Academic, 2017)
Jiang, H. et al. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell 137, 1343–1355 (2009)
Buchon, N. et al. Morphological and molecular characterization of adult midgut compartmentalization in Drosophila. Cell Rep. 3, 1725–1738 (2013)
Harrison, D. A. & Perrimon, N. Simple and efficient generation of marked clones in Drosophila. Curr. Biol. 3, 424–433 (1993)
Takeishi, A. et al. Homeostatic epithelial renewal in the gut is required for dampening a fatal systemic wound response in Drosophila. Cell Rep. 3, 919–930 (2013)
Hermiston, M. L. & Gordon, J. I. In vivo analysis of cadherin function in the mouse intestinal epithelium: essential roles in adhesion, maintenance of differentiation, and regulation of programmed cell death. J. Cell Biol. 129, 489–506 (1995)
Steinhusen, U. et al. Cleavage and shedding of E-cadherin after induction of apoptosis. J. Biol. Chem. 276, 4972–4980 (2001)
Maeda, K., Takemura, M., Umemori, M. & Adachi-Yamada, T. E-cadherin prolongs the moment for interaction between intestinal stem cell and its progenitor cell to ensure Notch signaling in adult Drosophila midgut. Genes Cells 13, 1219–1227 (2008)
Choi, N. H., Lucchetta, E. & Ohlstein, B. Nonautonomous regulation of Drosophila midgut stem cell proliferation by the insulin-signaling pathway. Proc. Natl Acad. Sci. USA 108, 18702–18707 (2011)
Biteau, B. & Jasper, H. EGF signaling regulates the proliferation of intestinal stem cells in Drosophila. Development 138, 1045–1055 (2011)
Buchon, N., Broderick, N. A., Kuraishi, T. & Lemaitre, B. Drosophila EGFR pathway coordinates stem cell proliferation and gut remodeling following infection. BMC Biol. 8, 152 (2010)
Jiang, H ., Grenley, M. O ., Bravo, M. J ., Blumhagen, R. Z . & Edgar, B. A. EGFR/Ras/MAPK signaling mediates adult midgut epithelial homeostasis and regeneration in Drosophila. Cell Stem Cell 8, 84–95 (2011)
Strand, M. & Micchelli, C. A. Regional control of Drosophila gut stem cell proliferation: EGF establishes GSSC proliferative set point & controls emergence from quiescence. PLoS ONE 8, e80608 (2013)
Qian, X., Karpova, T., Sheppard, A. M., McNally, J. & Lowy, D. R. E-cadherin-mediated adhesion inhibits ligand-dependent activation of diverse receptor tyrosine kinases. EMBO J. 23, 1739–1748 (2004)
Shilo, B. Z. Developmental roles of Rhomboid proteases. Semin. Cell Dev. Biol. 60, 5–9 (2016)
Zhou, F., Rasmussen, A., Lee, S. & Agaisse, H. The UPD3 cytokine couples environmental challenge and intestinal stem cell division through modulation of JAK/STAT signaling in the stem cell microenvironment. Dev. Biol. 373, 383–393 (2013)
Osman, D. et al. Autocrine and paracrine unpaired signaling regulate intestinal stem cell maintenance and division. J. Cell Sci. 125, 5944–5949 (2012)
McCrea, P. D. & Gottardi, C. J. Beyond β-catenin: prospects for a larger catenin network in the nucleus. Nat. Rev. Mol. Cell Biol. 17, 55–64 (2016)
Huang, J., Zhou, W., Dong, W., Watson, A. M. & Hong, Y. From the cover: directed, efficient, and versatile modifications of the Drosophila genome by genomic engineering. Proc. Natl Acad. Sci. USA 106, 8284–8289 (2009)
Chen, H., Zheng, X. & Zheng, Y. Age-associated loss of lamin-B leads to systemic inflammation and gut hyperplasia. Cell 159, 829–843 (2014)
Marianes, A. & Spradling, A. C. Physiological and stem cell compartmentalization within the Drosophila midgut. eLife 2, e00886 (2013)
Kolahgar, G. et al. Cell competition modifies adult stem cell and tissue population dynamics in a JAK–STAT-dependent manner. Dev. Cell 34, 297–309 (2015)
Lee, T . & Luo, L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22, 451–461 (1999)
Shaw, R. L. et al. The Hippo pathway regulates intestinal stem cell proliferation during Drosophila adult midgut regeneration. Development 137, 4147–4158 (2010)
Micchelli, C. A. & Perrimon, N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature 439, 475–479 (2006)
Ohlstein, B. & Spradling, A. The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature 439, 470–474 (2006)
O’Brien, L. E. Regional specificity in the Drosophila midgut: setting boundaries with stem cells. Cell Stem Cell 13, 375–376 (2013)
Rera, M. et al. Modulation of longevity and tissue homeostasis by the Drosophila PGC-1 homolog. Cell Metab. 14, 623–634 (2011)
Daniels, D. L. & Weis, W. I. β-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nat. Struct. Mol. Biol. 12, 364–371 (2005)
Zhang, T. & Du, W. Groucho restricts rhomboid expression and couples EGFR activation with R8 selection during Drosophila photoreceptor differentiation. Dev. Biol. 407, 246–255 (2015)
Acknowledgements
J.L. was supported by NSF GRFP DGE-114747 and NIH T32GM007276. This work was supported by NIH R03DK104027 and R01GM116000-01A1 to L.E.O. Confocal microscopy was performed at the Stanford Beckman Cell Sciences Imaging Facility (NIH 1S10OD01058001A1). We thank D. Bilder for the Caspase-3 antibody; the Developmental Studies Hybridoma Bank for other antibodies; D. Bilder, B. Edgar, M. Fuller, H. Jiang, B. Ohlstein, C. Thummel, A. Orian, the Bloomington Drosophila Stock Center (NIH P40OD018537), the TRiP at Harvard Medical School (NIH/NIGMS R01-GM084947) and the Vienna Drosophila Resource Center (http://stockcenter.vdrc.at/control/main) for fly stocks; J. Axelrod, M. Goodman, M. Fuller, W. J. Nelson, R. Nusse, M. Krasnow, T. Nystul and D. Fox for comments on the manuscript; and M. Mirvis, B. Benham-Pyle, N. Pierce and D. Gordon for helpful discussions.
Author information
Authors and Affiliations
Contributions
J.L. and L.E.O. designed all experiments and wrote the manuscript. J.L. and S.N. dissected and immunostained tissue samples. J.L., S.B. and S.N. performed confocal microscopy on tissue samples. J.L. performed qPCR assays, feeding assays and genetic crosses. J.L. also performed all quantitative analysis of microscopy data (for example, cell numbers, clone sizes, cell distances) and all statistical analysis.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks B. Edgar, R. Xi and Y. Yamashita for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
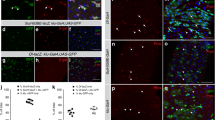
Extended Data Figure 1 Midgut lineage and morphology, esgF/O labelling system and workflow for semi-automated cell counting.
a, Lineage of the adult Drosophila midgut29,30. Stem cells are, in general, the only cells capable of division. Asymmetric stem cell divisions typically produce absorptive enterocytes; less frequently, they produce secretory enteroendocrine cells. Enterocytes arise through direct maturation of transient, post-mitotic intermediates called enteroblasts. Stem and enteroblast cells express the Snail-family transcription factor escargot (esg). b, Compartments of the female adult midgut7,25,31. The R4ab compartment (also known as P1–2)25 was used for all experiments in this study. Midgut schematic based on ref. 7. c–e, Identification of R4ab using morphological landmarks. As defined in ref. 7, R4ab is bounded by the apex of the midgut tube’s most distal 180° turn (blue arrowheads in d) and the first prominent muscle constriction distal to this 180° turn (red arrowheads in d). e, The R4ab distal muscle constriction (red arrowheads) is particularly apparent in confocal optical sections. Visceral muscle stained with phalloidin. Midguts in c, d and in e are two different samples. f, Genetic schema of the esgF/O system6. Stem and enteroblast cells are induced to express heritable GFP by temperature shift from 18 °C to 29 °C. The temperature shift inactivates GAL80ts, which allows the stem- and enteroblast-specific esgGAL4 to drive expression of both UAS-GFP and UAS-flp. The flp-recombinase renders GFP expression permanent and heritable by excising a CD2 ‘flp-out’ cassette to generate a functional actGAL4. Once generated, actGAL4 drives expression of UAS-GFP (and UAS-flp) irrespective of cell type. Therefore, after the temperature shift, all mature cells that arise from undifferentiated cells will express GFP. g, Pipeline for semi-automated, comprehensive cell counts of 3D-reconstructed midgut regions. (1) Confocal microscope z stacks capturing the entire depth of the organ are visualized in Fiji. (2) The R4ab region of the midgut (yellow outline) is digitally isolated and exported to Imaris. (Only the top half of the gut tube is shown.) Note that different midgut regions have different rates of turnover: R4ab undergoes complete turnover between adult days 4 and 8 (at 29 °C). However, other regions undergo slower turnover, as shown by large unlabelled regions outside of R4ab (see Methods). (3) To quantify total cells, nuclei (DAPI) are mapped to surface objects using Imaris. To quantify newly added cells in the esgF/O system, GFP+ cells are recognized in Imaris by co-localization of GFP and DAPI channels, and subsequently mapped to surface objects. Scale bars, 100 μm.
Extended Data Figure 2 Genetic schema of system to simultaneously manipulate enterocyte expression and trace stem cell divisions.
a, Detailed explanation of the genetic system in Fig. 1f. Flies are raised at 18 °C; at this temperature, GAL80ts represses mex-driven GAL4 in enterocytes (mexts) and lacZ labelling of stem cells is not induced. At four days after eclosion, flies are temperature-shifted to 29 °C; consequent inactivation of GAL80ts allows mexGAL4 to express genes of interest (UAS-gene X) specifically in enterocytes. After one day of UAS gene expression (five days after eclosion), flies are shifted to 38.5 °C for 1 h to induce ubiquitous expression of flp-recombinase, which is under control of a heat-shock promoter (hs-flp). Flp catalyses trans-recombination of the two FRTs to place the α-tubulin promoter upstream of the promoter-less nls-lacZ cassette and, consequently, turn on permanent nls-lacZ expression. After heat shock, flies are returned to 29 °C to maintain UAS-transgene expression. Midguts are collected for clonal analysis four days after the 38.5 °C heat shock (nine days after eclosion). See Methods. b, Validation of genetic system using mexts>his2av::RFP. β-Galactosidase marks a stem cell clone (outlined) in a background of His2av::RFP+ enterocytes. Within the 5-cell clone, only the enterocyte (yellow asterisk, polyploid) expresses his2av::RFP.
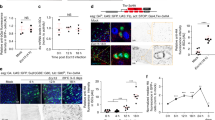
Extended Data Figure 3 Quantification of organ size and Egfr activation in genetically manipulated midguts.
a, Lengths of the R4ab compartment. n = 15, 11, 12, 10, 10, 11, 11, 11, 9, 10, 12, 11, 9, 10, 13, 11, 13, and 11 midguts from left to right, analysed after four days of UAS-transgene expression. b, dpErk+ cells in the R4ab compartment. n = 4, 4, 4, 5, 5, 4, 3, 4, 4, 3, 3, 4, 5, 3, 4, 3, 4, 4, and 4 midguts from left to right, analysed after two days of UAS-transgene expression. One of two replicate experiments was quantified. a, b, Box-and-whisker plots as in Fig. 1j, k; P values by unpaired t-test compared to control.
Extended Data Figure 4 Analysis of epithelial architecture, polarity and barrier function.
a–f, Apoptotic inhibition (b, e) or E-cad depletion (c, f) in enterocytes does not disrupt either epithelial architecture or apical–basal polarity. Images show vertical sections through the midgut epithelium after four days of either mexts>p35 or mexts>E-cad RNAi expression. Enterocytes remain as a coherent monolayer. Apical–basal polarity is intact, as revealed by immunolocalization of apical, actin-rich microvilli (a–f; SiR-Actin, red) and of two apico-lateral septate junction proteins, Coracle (a–c, green) and Discs-large (d–f, green). At the basal surface of the epithelium (white, dotted lines), midgut visceral muscle cells stain brightly for actin and Discs-large. Scale bars, 25 μm. g–j, Depletion of E-cad in enterocytes does not compromize the intestinal barrier. To test the intestinal barrier, flies were subjected to Smurf assays32 in which a blue, non-absorbable food dye is administered by feeding. The dye remains within the midgut (arrowhead) when the barrier is intact (g, non-Smurf), but leaks into the body cavity when the barrier is compromized, such as after the consumption of 1% SDS (h, Smurf). After 10 days of mexts>E-cad RNAi expression, midguts still retain the blue dye; no Smurf phenotypes were observed (i, j).
Extended Data Figure 5 Depletion of E-cad has distinct cell-autonomous and tissue-level effects on cell death.
In Fig. 2h, total cell counts show that midguts accumulate excess cells when E-cad is depleted from apoptosis-inhibited enterocytes, but not apoptosis-competent enterocytes. To shed light on this difference, we examined whether E-cad depletion itself promotes cell death. Two approaches, mosaic knockdown and pan-enterocyte knockdown, were used to distinguish direct, cell-autonomous effects from indirect, tissue-level effects. a–c, Mosaic knockdown of E-cad does not promote cell-autonomous death. To generate a mosaic epithelium, MARCM labelling27 was used to induce sparse, multicellular, GFP-marked clones in a background of unmarked, genetically unperturbed cells. a, b, Dotted outlines show representative control and E-cad RNAi clones (green). Sytox (red) identifies dying cells. c, Percentage of GFP+ cells that are also Sytox+. Dying cells occur with near-equal frequency within control and E-cad RNAi clones. Five midguts per genotype were analysed nine days after clone induction; n = 873 cells in control clones and 698 cells in E-cad RNAi clones. d–f, Pan-enterocyte knockdown of E-cad promotes cell death, probably through a non-autonomous effect. d, e, Representative images of mexts control and mexts>E-cad RNAi epithelia. Sytox (red) identifies dying cells. f, Quantification of Sytox+ cells in the R4ab compartment. The number of dying cells increases approximately 2.5× in E-cad RNAi midguts compared to control. n = 5 midguts per genotype, analysed after three days of transgene induction. c, f, Box-and-whisker plots as in Fig. 1j, k; P values by unpaired t-test. Scale bars, 25 μm. g, Summary. The unaltered frequency of dying cells in E-cad RNAi mosaic clones indicates that loss of E-cad does not cause cell-autonomous death. This result suggests that increased cell death in mexts>E-cad RNAi guts is a non-autonomous, tissue-level effect, possibly due to excess divisions (Fig. 2b) and consequent crowding. These findings may explain why p35, E-cad RNAi guts accumulate excess cells, whereas E-cad RNAi guts retain a normal number of cells (Fig. 2h).
Extended Data Figure 6 Loss of enterocyte E-cad activates Egfr, but not Wg, Hpo or Upd–Jak–Stat.
a, Effect of enterocyte E-cad depletion on target mRNA expression of known midgut regulatory pathways. mRNA expression was measured by qPCR of mexts control or mexts>E-cad RNAi midguts. Relative to control (dotted line), mRNA expression levels do not increase for the Wg targets frizzled-3 (fz3) and senseless (sens), the Hpo–Yki targets expanded (ex) and diap1, the injury-associated cytokines upd and upd3, and the Jak–Stat target windpipe (wdp). The other Jak–Stat target, Socs36E, is increased, which probably reflects its occasional activation in enterocytes (f). By comparison, the Egfr target pointed (pnt) is slightly increased and the Egfr target cyclinE (cycE) is substantially increased. Values are mean ± s.d.; data points are individual biological replicates analysed four days after induction. b–d, The number of upd3-lacZ+ enterocytes in the R4ab compartment is unchanged by enterocyte E-cad depletion. e–g, The number of 10×Stat-GFP+ diploid cells in R4ab is unchanged by enterocyte E-cad depletion. Occasional activation of 10×Stat-GFP+ occurs in E-cad-depleted enterocytes (asterisk), consistent with increased Socs36E (a). h–j, The number of cycE+ diploid cells in R4ab increases after enterocyte E-cad depletion. d, g, j, Box-and-whisker plots as in Fig. 1j, k; P values by unpaired t-test; NS, not significant. n = 4 midguts per condition, analysed two days after induction. One of two replicate experiments was quantified. k, dpErk immunostaining is limited to stem cells (Hrp+, Su(H)lacZ−; arrowheads) and does not mark enteroblasts (Hrp+, Su(H)lacZ+; asterisks), even in mexts>E-cad RNAi midguts. l, Expression of upd3 is not associated with physiological apoptosis. Most enterocytes that express upd3-lacZ are non-apoptotic, as assessed by staining for cleaved caspase-3. Values are means ± s.d.; n = 4 midguts per condition, analysed six days after eclosion. Representative images are shown in all panels. Scale bars, 25 μm.
Extended Data Figure 7 Two E-cad-associated transcription factors, Armadillo and p120-catenin, activate rho after loss of E-cad in enterocytes.
a, rho mRNA levels were measured by qPCR of mexts>E-cad RNAi midguts with additional manipulation of candidate rho regulators as indicated. Five candidates were examined: yki, a transcriptional co-activator in the Hpo pathway; groucho, a co-repressor known to target rho in some tissues33,34; puckered (puc), an inhibitor of Basket/Jnk, the latter of which can enhance Egf signalling; and arm and p120, co-activators that are inhibited by sequestration at E-cad adherens junctions. Knockdown of either arm or p120 significantly reduces rho activation. P values by unpaired t-test compared to E-cad RNAi alone. b, Overexpression of p120, but not armS10, in enterocytes is sufficient to increase rho mRNA levels. a, b, Values are mean ± s.d.; three biological replicates per condition, analysed four days after induction. Dotted lines show rho mRNA levels in unmanipulated controls. c–f, Depletion of E-cad or overexpression of p120 induces rho-lacZ in enterocytes and not in diploid cells. f, Quantification. g–l, Enterocyte arm and p120, but not yki or upd3, are necessary for activation of stem cell Egfr (dpErk immunostaining) following loss of E-cad. m–o, Enterocyte rho is necessary and sufficient for activation of stem cell Egfr. p–r, Enterocyte p120, but not arm, is sufficient to activate stem cell Egfr. See also Extended Data Fig. 3b. s, Overexpression of enterocyte rho increases the number of mitotic (phospho-histone H3+) stem cells. f, s, Box-and-whisker plots as in Fig. 1j, k; assessed after two days of transgene expression; P values by unpaired t-test compared to control; n = 5, 5, 5, 4 and 3 midguts from left to right (f); and n = 5 midguts (control) and 4 midguts (rho) (s). Representative images are shown in all panels. Scale bars, 25 μm.
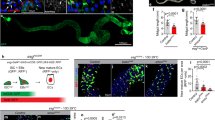
Extended Data Figure 8 Loss of rho, arm or p120 in enterocytes results in organ atrophy.
a, Total cell counts. Depletion of rho, arm or p120 in enterocytes reduces total cells compared to control. b, Depletion of enterocyte rho, arm or p120 reduces the length of the R4ab compartment compared to control. a, b, Box-and-whisker plots as in Fig. 1j, k; n = 5 midguts per genotype, analysed after six days of induction; P values by unpaired t-test compared to control. One of three independent experiments is shown in each graph. c, d, Depletion of enterocyte rho leads to organ atrophy. Representative whole mount images are shown. A, anterior; P, posterior. Scale bar, 200 μm.
Supplementary information
Supplementary Information
This file contains a Supplementary Discussion and Supplementary Tables 1-2 (full lists of experiment genotypes and primers used in this study).
Rights and permissions
About this article
Cite this article
Liang, J., Balachandra, S., Ngo, S. et al. Feedback regulation of steady-state epithelial turnover and organ size. Nature 548, 588–591 (2017). https://doi.org/10.1038/nature23678
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature23678
This article is cited by
-
Drosophila caspases as guardians of host-microbe interactions
Cell Death & Differentiation (2023)
-
A distinct Acyl-CoA binding protein (ACBP6) shapes tissue plasticity during nutrient adaptation in Drosophila
Nature Communications (2023)
-
Basal stem cell progeny establish their apical surface in a junctional niche during turnover of an adult barrier epithelium
Nature Cell Biology (2023)
-
Sexual identity of enterocytes regulates autophagy to determine intestinal health, lifespan and responses to rapamycin
Nature Aging (2022)
-
Long-lasting geroprotection from brief rapamycin treatment in early adulthood by persistently increased intestinal autophagy
Nature Aging (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.