Abstract
The type 2 cytokines interleukin (IL)-4, IL-5, IL-9 and IL-13 have important roles in stimulating innate and adaptive immune responses that are required for resistance to helminth infection, promotion of allergic inflammation, metabolic homeostasis and tissue repair1,2,3. Group 2 innate lymphoid cells (ILC2s) produce type 2 cytokines, and although advances have been made in understanding the cytokine milieu that promotes ILC2 responses4,5,6,7,8,9, how ILC2 responses are regulated by other stimuli remains poorly understood. Here we demonstrate that ILC2s in the mouse gastrointestinal tract co-localize with cholinergic neurons that express the neuropeptide neuromedin U (NMU)10,11. In contrast to other haematopoietic cells, ILC2s selectively express the NMU receptor 1 (NMUR1). In vitro stimulation of ILC2s with NMU induced rapid cell activation, proliferation, and secretion of the type 2 cytokines IL-5, IL-9 and IL-13 that was dependent on cell-intrinsic expression of NMUR1 and Gαq protein. In vivo administration of NMU triggered potent type 2 cytokine responses characterized by ILC2 activation, proliferation and eosinophil recruitment that was associated with accelerated expulsion of the gastrointestinal nematode Nippostrongylus brasiliensis or induction of lung inflammation. Conversely, worm burden was higher in Nmur1−/− mice than in control mice. Furthermore, use of gene-deficient mice and adoptive cell transfer experiments revealed that ILC2s were necessary and sufficient to mount NMU-elicited type 2 cytokine responses. Together, these data indicate that the NMU–NMUR1 neuronal signalling circuit provides a selective mechanism through which the enteric nervous system and innate immune system integrate to promote rapid type 2 cytokine responses that can induce anti-microbial, inflammatory and tissue-protective type 2 responses at mucosal sites.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Pulendran, B. & Artis, D. New paradigms in type 2 immunity. Science 337, 431–435 (2012)
Hammad, H. & Lambrecht, B. N. Barrier epithelial cells and the control of type 2 immunity. Immunity 43, 29–40 (2015)
Locksley, R. M. Asthma and allergic inflammation. Cell 140, 777–783 (2010)
Spits, H. et al. Innate lymphoid cells—a proposal for uniform nomenclature. Nat. Rev. Immunol. 13, 145–149 (2013)
Artis, D. & Spits, H. The biology of innate lymphoid cells. Nature 517, 293–301 (2015)
Robinette, M. L. et al. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat. Immunol. 16, 306–317 (2015)
Diefenbach, A., Colonna, M. & Koyasu, S. Development, differentiation, and diversity of innate lymphoid cells. Immunity 41, 354–365 (2014)
Walker, J. A., Barlow, J. L. & McKenzie, A. N. Innate lymphoid cells—how did we miss them? Nat. Rev. Immunol. 13, 75–87 (2013)
Klose, C. S. & Artis, D. Innate lymphoid cells as regulators of immunity, inflammation and tissue homeostasis. Nat. Immunol. 17, 765–774 (2016)
Howard, A. D. et al. Identification of receptors for neuromedin U and its role in feeding. Nature 406, 70–74 (2000)
Brighton, P. J., Szekeres, P. G. & Willars, G. B. Neuromedin U and its receptors: structure, function, and physiological roles. Pharmacol. Rev. 56, 231–248 (2004)
Veiga-Fernandes, H. & Mucida, D. Neuro-Immune Interactions at Barrier Surfaces. Cell 165, 801–811 (2016)
Gautron, L. et al. Neuronal and nonneuronal cholinergic structures in the mouse gastrointestinal tract and spleen. J. Comp. Neurol. 521, 3741–3767 (2013)
Hoyler, T. et al. The transcription factor GATA-3 controls cell fate and maintenance of type 2 innate lymphoid cells. Immunity 37, 634–648 (2012)
Moriyama, M. et al. The neuropeptide neuromedin U promotes inflammation by direct activation of mast cells. J. Exp. Med. 202, 217–224 (2005)
Hanada, R. et al. Neuromedin U has a novel anorexigenic effect independent of the leptin signaling pathway. Nat. Med. 10, 1067–1073 (2004)
Price, A. E. et al. Systemically dispersed innate IL-13-expressing cells in type 2 immunity. Proc. Natl Acad. Sci. USA 107, 11489–11494 (2010)
Halim, T. Y., Krauss, R. H., Sun, A. C. & Takei, F. Lung natural helper cells are a critical source of TH2 cell-type cytokines in protease allergen-induced airway inflammation. Immunity 36, 451–463 (2012)
Mjösberg, J. M. et al. Human IL-25- and IL-33-responsive type 2 innate lymphoid cells are defined by expression of CRTH2 and CD161. Nat. Immunol. 12, 1055–1062 (2011)
Monticelli, L. A. et al. Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nat. Immunol. 12, 1045–1054 (2011)
Schrage, R. et al. The experimental power of FR900359 to study Gq-regulated biological processes. Nat. Commun. 6, 10156 (2015)
Neill, D. R. et al. Nuocytes represent a new innate effector leukocyte that mediates type-2 immunity. Nature 464, 1367–1370 (2010)
Moro, K. et al. Innate production of TH2 cytokines by adipose tissue-associated c-Kit+Sca-1+ lymphoid cells. Nature 463, 540–544 (2010)
Moriyama, M. et al. The neuropeptide neuromedin U activates eosinophils and is involved in allergen-induced eosinophilia. Am. J. Physiol. Lung Cell. Mol. Physiol. 290, L971–L977 (2006)
Tallini, Y. N. et al. BAC transgenic mice express enhanced green fluorescent protein in central and peripheral cholinergic neurons. Physiol. Genomics 27, 391–397 (2006)
Rossi, J. et al. Melanocortin-4 receptors expressed by cholinergic neurons regulate energy balance and glucose homeostasis. Cell Metab. 13, 195–204 (2011)
Madisen, L. et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010)
Rawlins, E. L., Clark, C. P., Xue, Y. & Hogan, B. L. The Id2+ distal tip lung epithelium contains individual multipotent embryonic progenitor cells. Development 136, 3741–3745 (2009)
Mohrs, M., Shinkai, K., Mohrs, K. & Locksley, R. M. Analysis of type 2 immunity in vivo with a bicistronic IL-4 reporter. Immunity 15, 303–311 (2001)
Yu, C. et al. Targeted deletion of a high-affinity GATA-binding site in the GATA-1 promoter leads to selective loss of the eosinophil lineage in vivo. J. Exp. Med. 195, 1387–1395 (2002)
Feyerabend, T. B. et al. Cre-mediated cell ablation contests mast cell contribution in models of antibody- and T-cell-mediated autoimmunity. Immunity 35, 832–844 (2011)
Hsu, C. L., Neilsen, C. V. & Bryce, P. J. IL-33 is produced by mast cells and regulates IgE-dependent inflammation. PLoS One 5, e11944 (2010)
Dodt, M., Roehr, J. T., Ahmed, R. & Dieterich, C. FLEXBAR-flexible barcode and adapter processing for next-generation sequencing platforms. Biology (Basel) 1, 895–905 (2012)
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013)
Liao, Y., Smyth, G. K. & Shi, W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41, e108 (2013)
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014)
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015)
Chung, K. et al. Structural and molecular interrogation of intact biological systems. Nature 497, 332–337 (2013)
Gabanyi, I. et al. Neuro-immune Interactions Drive Tissue Programming in Intestinal Macrophages. Cell 164, 378–391 (2016)
Tait Wojno, E. D. et al. The prostaglandin D2 receptor CRTH2 regulates accumulation of group 2 innate lymphoid cells in the inflamed lung. Mucosal Immunol. 8, 1313–1323 (2015)
Acknowledgements
We thank I. Gabanyi and D. Mucida for help with the muscularis isolation, H.-R. Rodewald for providing Cpa3cre and G. Eberl for RorgtGFP mice. We thank the Epigenomics Core, the Imaging Core and the Mouse Genetics Core at Weill Cornell Medicine and MSKCC. Nmur1LacZ/+ mice were generated by Velocigene and NmuGFP by GENSAT and provided by the KOMP or MMRRC Repository at UC Davis. The work was supported by grants from the German Research Foundation (DFG; KL 2963/1-1 to C.S.N.K.; FOR2372 to E.K. and G.M.K.), the Australian National Health and Medical Research Comission (NHMRC) early career fellowship (to L.C.R.), the Novo Nordic Foundation (14052; to J.B.M.), the Weill Cornell Department of Medicine Pre-Career Award (to L.A.M.), the Naito Foundation (to S.M.), JSPS Overseas Research Fellowships (to S.M.), MSD Life Science Foundation (to H.K.), the Jill Roberts Institute (to G.G.P.), Defense Advanced Research Projects Agency (DARPA; HR0011-16-C-0138 to X.S.), the National Institutes of Health (NIH; F32AI134018 to L.A.M.; AI061570, AI087990, AI074878, AI083480, AI095466, AI095608, AI102942, AI106697 and AI097333 to D.A.; R01GM114254 and OT2-OD023849 to X.S.; AI106697 to D.L.F.), the Burroughs Wellcome Fund (to D.A.) and the Crohn’s & Colitis Foundation of America (to T.M. and D.A.).
Author information
Authors and Affiliations
Contributions
C.S.N.K. carried out most experiments and analysed the data. T.M., J.B.M., L.C.R., A.-L.F., H.K., L.A.M., S.M., N.R. and X.S. helped with experiments, and E.K. and G.M.K. provided the inhibitor FR900359 (previous commercial name UBO-QIC). G.G.P. performed RNA-seq analysis. T.S., D.C. and D.L.F. provided human tissue samples. D.A. and C.S.N.K. conceived the project, analysed data, and wrote the manuscript with input from all co-authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks W. J. de Jonge and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 ILC2s and neurons co-localize.
Surface reconstruction of immunofluorescence staining from the intestinal submucosa shown in Fig. 1a. Scale bar, 30 μm. Data are representative of three independent experiments with similar results.
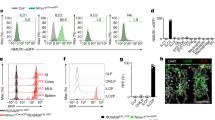
Extended Data Figure 2 ILC2s selectively express NMUR1.
a, Expression of Nmur1 in the indicated sorted cell populations as determined by qPCR analysis (n = 5). MC, mast cells; MΦ, macrophages; PMΦ, peritoneal macrophages. MC and PMΦ were obtained by peritoneal lavage; MΦ and ILC2s were purified from the small intestine. b–d, f, Histograms and dot plots show expression of Nmur1 as measured by conversion of the fluorescent LacZ substrate FDG. Histograms are gated on Lin−CD45+ cells and CD127+KLRG1+ (ILC2s), CD127+CCR6+ (ILC3s), CD127+CCR6−NKp46+NK1.1+ (ILC1s), CD3+ (T cells), CD19+ (B cells), CD3−CD19−CD11b+SiglecF+ (Eosinophils) from the small intestine (b, c). Gating for mast cells and basophils from the lung is shown (d). Percentage (n = 3) of FDG+ cells from the indicated population of the small intestine. Eos, eosinophils (f). e, g, Flow cytometry analysis of the indicated immune cell populations (top row) for Nmur1 (bottom row). MC and PMΦ were obtained by peritoneal lavage; MΦ and ILC2 were purified from the small intestine (e). Percentage (n = 3) of FDG+ cells (g). h, Expression of Nmu (n = 3 (LPL), n = 5 (all others)), as determined by qPCR from the indicated fractions of the murine small intestine. i, Expression of NMU (n = 9 (epithelium), n = 8 (whole jejunum and LPL), n = 7 (IEL), n = 6 (parenchyma)) as determined by qPCR from the indicated fractions of the human jejunum. j, k, CLARITY staining of the small intestine (j) or colon (k) for NMU. l, Image of the intestinal muscularis mucosae from NmuGFP mice. m, Immunofluorescence staining of the intestinal mucosa from Chatcre × Ai14 mice for NMU. Scale bar, 100 μm (j–m). Error bars, mean + s.d. Data are representative of two (e, g) or three independent experiments (b–f, j–m) with similar results. Data in a, h, i are based on the indicated number of biological replicates per group.
Extended Data Figure 3 ILC2s and NMU+ neurons co-localize.
a, Surface reconstruction of immunofluorescent staining from the intestinal submucosa shown Fig. 1i. Scale bar, 50 μm. b, Percentage (n = 4) of ILC2s, that have overlapping pixels with neurons. A total of 348 cells were counted and 236 cells exhibited pixels overlapping with NMU staining. c, Expression of Nmu as determined by qPCR in enteric neuron (n = 5, pooled from two independent experiments) cultures and compared to the epithelial fraction or parenchyma of the small intestine (n = 3). d, e, Sort-purified ILC2s (3 × 104) were cultured with or without enteric neurons for 5 days. Absolute number (d) and FSC (e) (n = 3) of ILC2s are shown. Error bars, mean + s.d. Data are representative of three independent experiments (a, d, e) or on the indicated number of biological replicates per group (b, c).
Extended Data Figure 4 NMU activates ILC2s.
a, Gating strategy for flow cytometric analysis of bulk LPLs cytokine assays. Lineage 1: CD11b, CD11c and B220 (all APC-eF780); lineage 2: CD3, CD5 (both PerCP-Cy5.5) and FcεRI PerCP-eF710. b, c, Concentration (n = 3) of IL-13 (b) or IL-5 (c) in the culture supernatant after 4 h stimulation of bulk LPLs with a control peptide or NMU as determined by ELISA (n.d., not detectable). d, Bulk LPLs from Il1rl1+/+ and Il1rl1−/− mice were incubated in medium with or without NMU for 4 h in vitro. Percentage (Il1rl1+/+, n = 5 and Il1rl1−/−, n = 4) of IL-5+KLRG1+ cells. e, f, LPLs from Nmur1+/+ or Nmur1−/− mice were analysed by flow cytometry. Plots are gated on Lin−CD45+ lymphocytes (e). Percentage (n = 3) of GATA-3+KLRG1+ cells (f). g, Bulk LPLs were incubated for 30 min with DMSO or the inhibitor of Gαq proteins FR900359 in vitro. Medium, NMU or the indicated cytokine cocktail was then added and the assay was incubated for another 4 h. Percentage (n = 3) of IL-13 YFP+KLRG1+ cells among all KLRG1+ cells. h, Percentage (n = 4) of IL-5+KLRG1+ or IL-13+KLRG1+ cells as determined by intracellular cytokine staining. i–k, Overnight incubation of sort-purified intestinal ILC2s from Il13YFP/+ mice in medium with or without NMU. FSC (n = 3) (i), histogram overlay of IL-13 YFP (j) and percentage (n = 3) of IL-13 YFP+ ILC2s (k). l, ILC2s from the small intestine were sort-purified and incubated in medium without or with NMU overnight in vitro. Contour plots show intracellular flow cytometry analysis for IL-5 and IL-13. Error bars, mean + s.d. Data are representative of two (b–d, g, h) or three independent experiments (e, f, i–l) with similar results. Gating in a is representative for cytokine assays used in the whole study.
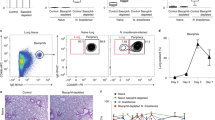
Extended Data Figure 5 Nmu stimulates ILC2s in vivo.
a, PBS or NMU was injected daily in C57BL/6 mice. After two days, ILC3s from the small intestine were analysed by flow cytometry for Ki67. Plots are gated on Lin−CD45+RORγt+ lymphocytes. Percentage (n = 3) of Ki67+ cells. b, c, PBS or NMU was injected daily for two days in CD45.1:Nmur1+/+/CD45.2:Nmur1−/− mixed bone marrow chimaeras. One day later, ILC2s from the small intestine were analysed by flow cytometry for KLRG1 and Ki67. Plots are gated on Lin−CD127+KLRG1+ lymphocytes and either CD45.1 or CD45.2 (b). KLRG1 mean fluorescence intensity (PBS n = 4, NMU n = 5) (c). d, PBS or NMU (100 μg) was injected in CD45.1:Nmur1+/+/CD45.2:Nmur1−/− mixed bone marrow chimaeras. One day later, ILC2s from the small intestine were analysed by flow cytometry for IL-5 and IL-13 expression. Percentage (n = 7 (PBS), n = 8 (NMU)) of IL-5+ and IL-13+ ILC2s. Plots are gated on Lin−CD127+KLRG1+ lymphocytes and either CD45.1 or CD45.2. e, PBS or NMU was injected once in Il13YFP/+ mice. FSC (n = 3) of Lin−CD45+CD127+CD25+KLRG1+ LPLs one day after injection. f, C57BL/6 mice were infected with T. muris (n = 11) or left untreated (n = 6). On day 18, Nmu expression was determined by qPCR in a piece of the proximal colon. g, C57BL/6 mice were infected with H. polygyrus (n = 18). Control C57BL/6 (n = 4) mice were left untreated. On day 18, Nmu expression was determined by qPCR in a piece of the duodenum. h, Nmur1+/+ or Nmur1LacZ/+ mice were infected with N. brasiliensis. Control Nmur1+/+ mice were left untreated. On day 7, mice (n = 6) were analysed. Histogram overlay shows expression of Nmur1 (FDG) on ILC2s from the small intestine and are gated on Lin−CD45+KLRG1+ lymphocytes. i, Nmur1+/+ or Nmur1LacZ/+ mice were infected with N. brasiliensis. Control mice were left untreated. On day 7, mice were analysed and the percentage of Nmur1+ (FDG) determined by flow cytometry in the indicated subsets. Percentage (n = 6 (intestinal subsets) or 8 (lung subsets) for infected Nmur1LacZ/+ and n = 3 for control mice) of Nmur1+ (FDG) cells. Plots are gated on CD45+ cells and FcεRI+CD49b+c-Kit− for basophils (BF), FcεRI+CD49b+c-Kit+ for mast cells, CD11b+F4/80+ for MΦ, CD3+CD5+ for T cells and Lin−KLRG1+ for ILC2s. j, Nmur1LacZ/+ mice were infected with N. brasiliensis. On day 14, mice were analysed and the percentage (n = 7 or 9 (lung)) of Nmur1+ (FDG) CD3+CD5+ T cells or Lin−KLRG1+ ILC2s was determined by flow cytometry. k, l, Il4GFP mice were infected with N. brasiliensis. On day 14, CD4+ T cells (gated on CD3+CD5+ lymphocytes) were sort-purified in IL-4-positive and -negative populations based on GFP expression (k) and Nmur1 expression was determined by qPCR (l) (n = 3 (ILC2), n = 4 (IL-4+CD4+ lung) or n = 5). m, n, PBS or NMU was injected daily for two days in C57BL/6 or Cpa3cre mice. One day later, ILC2s from the small intestine were analysed by flow cytometry for KLRG1 and Ki67 expression. Plots are gated on Lin−CD45+GATA-3+KLRG1+ cells (m). Percentage (n = 5 or 3 (PBS)) of Ki67+KLRG1+ cells among all KLRG1+ cells (n). o, PBS or NMU was injected daily for two days in BALB/c or ΔdblGATA1 mice. One day later, ILC2s from the small intestine were analysed by flow cytometry for KLRG1 and Ki67 expression. Percentage (n = 6 or 5 (PBS ΔdblGATA1)) of Ki67+KLRG1+ cells among all KLRG1+ cells. p, Bone marrow chimaeras reconstituted with Nmur1+/+ or Nmur1−/− bone marrow were infected subcutaneously with N. brasiliensis. On day 7, worm burden (n = 15 (Nmur1+/+), n = 14 (Nmur1−/−)) in the small intestine was quantified. q, Rag2−/−Il2rg−/− mice were reconstituted with ILC2 precursors from Nmur1+/+ or Nmur1−/− mice. After reconstitution, mice were infected with N. brasiliensis and NMU (20 μg) was injected i.p. on day 2, 4 and 6. Plots show flow cytometry analysis of cells from the lung and are gated on CD45+CD11c− cells. Error bars, mean + s.d. Data are representative of two (b, c, k–o, q) or three independent experiments (a, e, h) with similar results. The data are pooled from two (d) or three (f, g, i) independent experiments. The data in j and p are representative of the indicated number of biological replicates per group.
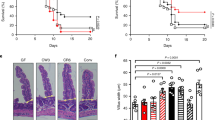
Extended Data Figure 6 ILC2 are required for NMU-induced lung inflammation.
a, PBS or NMU was intranasally administered to C57BL/6 mice daily for four days. One day later, ILC2s from the lung were analysed by flow cytometry. Plots are gated on Lin–CD45+GATA-3+CD25+ lymphocytes. b, PBS or NMU was delivered intranasally to C57BL/6 mice daily for five days. Three days later, eosinophil infiltration was determined in BAL by flow cytometry. Percentage (mean + s.d., n = 5) of CD11b+SiglecF+CD11c– eosinophils in the BAL. c, PBS or NMU was intranasally administered to Rag2−/− or Rag2−/−Il2rg−/− mice daily for four days. One day later, ILC2s and eosinophils from the lung were analysed by flow cytometry. d, PBS or NMU was intranasally administered daily for four days to Rag2−/−Il2rg−/− mice or Rag2−/−Il2rg−/− that were reconstituted with ILC2 progenitors. One day later, ILC2s and eosinophils from the lung were analysed by flow cytometry. e, PBS or NMU was intranasally administered to Nmur1+/+ or Nmur1−/− mice daily for four days. One day later, ILC2s and eosinophils from the lung were analysed by flow cytometry. Plots are gated on Lin−CD45+CD25+GATA-3+ lymphocytes. Data are representative of two (d, e) or three independent experiments (a–c) with similar results.
Supplementary information
Rights and permissions
About this article
Cite this article
Klose, C., Mahlakõiv, T., Moeller, J. et al. The neuropeptide neuromedin U stimulates innate lymphoid cells and type 2 inflammation. Nature 549, 282–286 (2017). https://doi.org/10.1038/nature23676
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature23676
This article is cited by
-
Cooperation of ILC2s and TH2 cells in the expulsion of intestinal helminth parasites
Nature Reviews Immunology (2024)
-
Nr4a1 marks a distinctive ILC2 activation subset in the mouse inflammatory lung
BMC Biology (2023)
-
Group 2 innate lymphoid cells and their surrounding environment
Inflammation and Regeneration (2023)
-
Role of thymic stromal lymphopoietin in allergy and beyond
Nature Reviews Immunology (2023)
-
Type 2 immunity in the brain and brain borders
Cellular & Molecular Immunology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.