Abstract
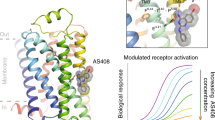
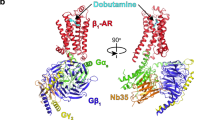
G-protein-coupled receptors (GPCRs) pose challenges for drug discovery efforts because of the high degree of structural homology in the orthosteric pocket, particularly for GPCRs within a single subfamily, such as the nine adrenergic receptors. Allosteric ligands may bind to less-conserved regions of these receptors and therefore are more likely to be selective. Unlike orthosteric ligands, which tonically activate or inhibit signalling, allosteric ligands modulate physiologic responses to hormones and neurotransmitters, and may therefore have fewer adverse effects. The majority of GPCR crystal structures published to date were obtained with receptors bound to orthosteric antagonists, and only a few structures bound to allosteric ligands have been reported. Compound 15 (Cmpd-15) is an allosteric modulator of the β2 adrenergic receptor (β2AR) that was recently isolated from a DNA-encoded small-molecule library1. Orthosteric β-adrenergic receptor antagonists, known as beta-blockers, are amongst the most prescribed drugs in the world and Cmpd-15 is the first allosteric beta-blocker. Cmpd-15 exhibits negative cooperativity with agonists and positive cooperativity with inverse agonists. Here we present the structure of the β2AR bound to a polyethylene glycol-carboxylic acid derivative (Cmpd-15PA) of this modulator. Cmpd-15PA binds to a pocket formed primarily by the cytoplasmic ends of transmembrane segments 1, 2, 6 and 7 as well as intracellular loop 1 and helix 8. A comparison of this structure with inactive- and active-state structures of the β2AR reveals the mechanism by which Cmpd-15 modulates agonist binding affinity and signalling.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Ahn, S. et al. Allosteric ‘beta-blocker’ isolated from a DNA-encoded small molecule library. Proc. Natl Acad. Sci. 114, 1708–1713 (2017)
Kruse, A. C. et al. Activation and allosteric modulation of a muscarinic acetylcholine receptor. Nature 504, 101–106 (2013)
Zheng, Y. et al. Structure of CC chemokine receptor 2 with orthosteric and allosteric antagonists. Nature 540, 458–461 (2016)
Oswald, C. et al. Intracellular allosteric antagonism of the CCR9 receptor. Nature 540, 462–465 (2016)
Jazayeri, A. et al. Extra-helical binding site of a glucagon receptor antagonist. Nature 533, 274–277 (2016)
Rosenbaum, D. M. et al. GPCR engineering yields high-resolution structural insights into β2-adrenergic receptor function. Science 318, 1266–1273 (2007)
Rasmussen, S. G. et al. Crystal structure of the β2 adrenergic receptor–Gs protein complex. Nature 477, 549–555 (2011)
Kang, Y. et al. Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature 523, 561–567 (2015)
Staus, D. P. et al. Allosteric nanobodies reveal the dynamic range and diverse mechanisms of G-protein-coupled receptor activation. Nature 535, 448–452 (2016)
Ring, A. M. et al. Adrenaline-activated structure of β2-adrenoceptor stabilized by an engineered nanobody. Nature 502, 575–579 (2013)
Rasmussen, S. G. et al. Structure of a nanobody-stabilized active state of the β2 adrenoceptor. Nature 469, 175–180 (2011)
Huang, W. et al. Structural insights into μ-opioid receptor activation. Nature 524, 315–321 (2015)
Kobilka, B. K. Amino and carboxyl terminal modifications to facilitate the production and purification of a G protein-coupled receptor. Anal. Biochem. 231, 269–271 (1995)
Caffrey, M. & Cherezov, V. Crystallizing membrane proteins using lipidic mesophases. Nat. Protoc. 4, 706–731 (2009)
Kabsch, W. Xds. Acta Crystallogr. D 66, 125–132 (2010)
Strong, M. et al. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc. Natl Acad. Sci. 103, 8060–8065 (2006)
Adams, P. D . et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010)
Ten Eyck, L. F. Fast Fourier transform calculation of electron density maps. Methods Enzymol. 115, 324–337 (1985)
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D 67, 235–242 (2011)
Krissinel, E. Stock-based detection of protein oligomeric states in jsPISA. Nucleic Acids Res. 43 (W1), W314–W319 (2015)
Whorton, M. R. et al. A monomeric G protein-coupled receptor isolated in a high-density lipoprotein particle efficiently activates its G protein. Proc. Natl Acad. Sci. 104, 7682–7687 (2007)
Strachan, R. T. et al. Divergent transducer-specific molecular efficacies generate biased agonism at a G protein-coupled receptor (GPCR). J. Biol. Chem. 289, 14211–14224 (2014)
Ghanouni, P. et al. The effect of pH on β2 adrenoceptor function. Evidence for protonation-dependent activation. J. Biol. Chem. 275, 3121–3127 (2000)
Ranganathan, A., Dror, R. O. & Carlsson, J. Insights into the role of Asp792.50 in β2 adrenergic receptor activation from molecular dynamics simulations. Biochemistry 53, 7283–7296 (2014)
Lomize, M. A., Lomize, A. L., Pogozheva, I. D. & Mosberg, H. I. OPM: orientations of proteins in membranes database. Bioinformatics 22, 623–625 (2006)
Huang, J. & MacKerell, A. D., Jr. CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 34, 2135–2145 (2013)
Klauda, J. B. et al. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B 114, 7830–7843 (2010)
MacKerell, A. D. et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 102, 3586–3616 (1998)
Best, R. B. et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ1 and χ2 dihedral angles. J. Chem. Theory Comput. 8, 3257–3273 (2012)
Best, R. B., Mittal, J., Feig, M. & MacKerell, A. D., Jr. Inclusion of many-body effects in the additive CHARMM protein CMAP potential results in enhanced cooperativity of α-helix and β-hairpin formation. Biophys. J. 103, 1045–1051 (2012)
Vanommeslaeghe, K. et al. CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 31, 671–690 (2010)
Vanommeslaeghe, K., Raman, E. P. & MacKerell, A. D., Jr. Automation of the CHARMM General Force Field (CGenFF) II: assignment of bonded parameters and partial atomic charges. J. Chem. Inf. Model. 52, 3155–3168 (2012)
Vanommeslaeghe, K. & MacKerell, A. D., Jr. Automation of the CHARMM General Force Field (CGenFF) I: bond perception and atom typing. J. Chem. Inf. Model. 52, 3144–3154 (2012)
Salomon-Ferrer, R., Götz, A. W., Poole, D., Le Grand, S. & Walker, R. C. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh ewald. J. Chem. Theory Comput. 9, 3878–3888 (2013)
Case, D. A. et al. AMBER 2016. (2016)
Hopkins, C. W., Le Grand, S., Walker, R. C. & Roitberg, A. E. Long-time-step molecular dynamics through hydrogen mass repartitioning. J. Chem. Theory Comput. 11, 1864–1874 (2015)
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996)
Wallace, A. C., Laskowski, R. A. & Thornton, J. M. LIGPLOT: a program to generate schematic diagrams of protein–ligand interactions. Protein Eng. 8, 127–134 (1995)
Acknowledgements
We thank K. Hirata at Beamline BL32XU of Spring-8 for assistance in data collection. A. Wall and T. Xu provided technical assistance. NuEvolution for constructive discussions in the course of the work. We acknowledge support from the National Institute of Health grants NS028471 and GM106990 (B.K.K.), HL16037 (R.J.L.) and T32HL007101 (A.W.K. and A.M.), Amgen-China Postdoc fellowship (X.L.) and the Mathers Foundation (B.K.K. and W.I.W.). R.J.L. is an investigator with the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
X.L. expressed and purified β2AR–T4L, performed crystallization, data collection, data processing, structure determination and refinement. S.A. designed and performed in vitro radio ligand binding and mutagenesis studies. A.W.K. designed the chemical synthetic route for Cmpd-15PA. B.P. designed and performed the in vitro radio ligand binding experiments. A.M. assisted in the design of mutagenesis studies and the development of Cmpd-15PA. K.-C.M. synthesized Cmpd-15PA and analysed the spectral data. X.C. designed the chemical structure and synthetic route for Cmpd-15PA, analysed the spectral data and assisted in manuscript preparation. N.R.L. performed and analysed molecular dynamics simulations, with assistance from A.J.V. R.O.D. oversaw molecular dynamics simulations and analysis. W.I.W. oversaw data processing, structure determination and refinement. The manuscript was written by B.K.K. with assistance from X.L. R.J.L. and B.K.K. coordinated the experiments and supervised the overall research. All authors contributed to the editing of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks T. Sakmar, P. Scheerer and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 Effects of Cmpd-15PA on agonist binding to the β2AR.
a, The level of 3H-Fenoterol (Fen) high-affinity binding to the β2AR was measured after pretreatment with the vehicle control (0.5% DMSO), Cmpd-15 or Cmpd-15PA at various concentrations as indicated in the presence of transducers, either trimeric Gαβγ protein or β-arrestin 1 (β-arr1) together with Fab30. Values were expressed as percentages of the maximal 3H-Fen binding level promoted by each transducer in the vehicle control (0.5% DMSO) and represent mean ± s.e.m. obtained from four independent experiments done in duplicate. b, Isoproterenol (ISO)-125I-CYP competition binding curves were obtained using wild-type (WT) β2AR or β2AR–T4L (T4L) in the absence (vehicle alone) or the presence of Cmpd-15PA at 32 μM. Values are expressed as percentages of the maximal 125I-CYP binding level obtained from a one-site competition binding-log IC50 curve fit and represent mean ± s.e.m. obtained from four independent experiments done in duplicate.
Extended Data Figure 2 Bromine atom of Cmpd-15PA was sensitive to radiation damage.
a–c, Clear differences were observed around the bromine atom of Cmpd-15PA between simulated annealing omit maps of the first half of diffraction data (a, green density, 2.3σ) and maps of the second half of diffraction data (b, green density, 2.3σ), resulting in a strong difference electron density centred around the bromine atom (c, purple density, 4σ).
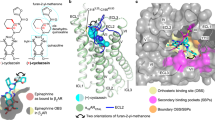
Extended Data Figure 3 Two-dimensional schematic depiction of the Cmpd-15PA binding pocket.
Carbon atoms are coloured in black, oxygen atoms in red, nitrogen atoms in blue, bromine atom in green. Hydrogen bonds are represented as green dashed lines with distances labelled. The figure is generated using LIGPLOT40.
Extended Data Figure 4 Crystallographic evidence support modelling of R isomer of Cmpd-15PA.
a, b, The R isomer fits better into the simulated annealing omit map (green density map, 2.3σ) than S isomer (b). c, When refined with R isomer, the Cmpd-15PA model fits well into the 2Fo − Fc density (grey map, 1.5σ) with only very weak negative Fo − Fc density (red map, 3.0σ). d, When refined with S isomer, the Cmpd-15PA model does not fit as well to the 2Fo − Fc density (grey map, 1.5σ). The negative density Fo − Fc map (red map, 3.0σ) and positive density Fo − Fc map (green map, 3.0σ) suggest that the S isomer is not supported by crystallographic data.
Extended Data Figure 5 Comparison of modulator binding pocket before and after Cmpd-15PA binding.
a, b, The binding pocket is occluded before Cmpd-15PA binding (a), which would be an obstacle for in silico docking because the shape of the pocket is markedly different after Cmpd-15 binding (b).
Extended Data Figure 6 Comparison of the structures of the β2AR (cyan, PDB, 2RH1), β2AR–Nb60 (green, PDB, 5JQH) and β2AR–Cmpd-15PA (orange).
a, Cmpd-15PA binding pocket overlaps with Nb60 binding pocket. b–d, Different views of superimposed structures of the β2AR, the β2AR–Nb60 and the β2AR–Cmpd-15PA revealing very little structural difference associated with binding of Nb60 or Cmpd-15PA.
Extended Data Figure 7 Relative activities of Cmpd-15 analogues in β2AR-mediated responses.
For each analogue, only the modified region relative to the parent Cmpd-15 is indicated. Values for ‘inhibition Emax (%)’ are expressed as percentages of analogue-induced inhibition relative the Cmpd-15-induced level. Values for ‘EC50 shift (fold)’ are expressed as rightward fold-shifts compared to the EC50 value obtained in the vehicle (DMSO)-treated control curve. All values represent mean ± s.e.m. obtained from at least four independent experiments done in duplicate. Statistical analyses were performed using ‘one-way ANOVA’ with ‘Dunnetts’ post-tests for comparison to the control. ***P < 0.001. cAMP, G protein-mediated cAMP accumulation; βarr, β-arrestin recruitment to the β2AR.
Extended Data Figure 8 Alignment of residues that form the Cmpd-15PA binding pocket in β2AR with those from β1AR, V2R and AT1R.
There are 21 residues from β2AR that form the Cmpd-15 binding pocket (highlighted green). The identical residues from β1AR, V2R and AT1R are also highlighted in green. 10 out of the 21 residues are located at TM1, ICL1 and TM2 (a), while 11 of the 21 residues are located at TM6, TM7 and helix 8 (b). The top numbering refers to protein sequences in β2AR. The bottom numbering refers to Ballesteros–Weinstein numbering. c, Cmpd-15 has no effect on orthosteric agonist binding to the β1AR. Dose–response curves of isoproterenol (ISO)-competition binding to the β1AR with 125I-CYP were obtained in the presence of various concentrations of Cmpd-15 as indicated. Values were expressed as percentages of the maximal 125I-CYP binding level obtained from a one-site competition binding-log IC50 curve fit and represent mean ± s.e.m. obtained from five independent experiments done in duplicate.
Extended Data Figure 9 Comparison of Cmpd-15PA pocket with intracellular allosteric antagonist pockets of CCR2 and CCR9.
a, Cmpd-15PA pocket is formed by residues from TM1, TM2, TM6, TM7, ICL1 and helix 8 in β2AR. b, c, The binding pocket of CCR2-RA-(R) in CCR2 (b) and the pocket of vercirnon in CCR9 (c) are similar to Cmpd-15PA (blue lines) pocket but involve more interactions with TM3. (Protein Data Bank accession numbers, 5T1A for CCR2/CCE2-RA-(R) and 5LWE for CCR9/vercirnon.)
Supplementary information
Supplementary Information
This file contains supplementary text and figures S1-S6. (PDF 1499 kb)
Rights and permissions
About this article
Cite this article
Liu, X., Ahn, S., Kahsai, A. et al. Mechanism of intracellular allosteric β2AR antagonist revealed by X-ray crystal structure. Nature 548, 480–484 (2017). https://doi.org/10.1038/nature23652
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature23652
This article is cited by
-
G protein-coupled receptors (GPCRs): advances in structures, mechanisms, and drug discovery
Signal Transduction and Targeted Therapy (2024)
-
A framework for Frizzled-G protein coupling and implications to the PCP signaling pathways
Cell Discovery (2024)
-
Function and dynamics of the intrinsically disordered carboxyl terminus of β2 adrenergic receptor
Nature Communications (2023)
-
Class B1 GPCR activation by an intracellular agonist
Nature (2023)
-
Small-molecule discovery through DNA-encoded libraries
Nature Reviews Drug Discovery (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.