Abstract
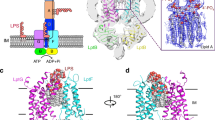
Lipopolysaccharide (LPS) in the outer membrane of Gram-negative bacteria is critical for the assembly of their cell envelopes. LPS synthesized in the cytoplasmic leaflet of the inner membrane is flipped to the periplasmic leaflet by MsbA, an ATP-binding cassette transporter. Despite substantial efforts, the structural mechanisms underlying MsbA-driven LPS flipping remain elusive. Here we use single-particle cryo-electron microscopy to elucidate the structures of lipid-nanodisc-embedded MsbA in three functional states. The 4.2 Å-resolution structure of the transmembrane domains of nucleotide-free MsbA reveals that LPS binds deep inside MsbA at the height of the periplasmic leaflet, establishing extensive hydrophilic and hydrophobic interactions with MsbA. Two sub-nanometre-resolution structures of MsbA with ADP-vanadate and ADP reveal an unprecedented closed and an inward-facing conformation, respectively. Our study uncovers the structural basis for LPS recognition, delineates the conformational transitions of MsbA to flip LPS, and paves the way for structural characterization of other lipid flippases.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Dowhan, W. Molecular basis for membrane phospholipid diversity: why are there so many lipids? Annu. Rev. Biochem. 66, 199–232 (1997)
Holthuis, J. C. & Menon, A. K. Lipid landscapes and pipelines in membrane homeostasis. Nature 510, 48–57 (2014)
van Meer, G. Dynamic transbilayer lipid asymmetry. Cold Spring Harb. Perspect. Biol. 3, a004671 (2011)
Sharom, F. J. Flipping and flopping–lipids on the move. IUBMB Life 63, 736–746 (2011)
Sebastian, T. T., Baldridge, R. D., Xu, P. & Graham, T. R. Phospholipid flippases: building asymmetric membranes and transport vesicles. Biochim. Biophys. Acta 1821, 1068–1077 (2012)
Andersen, J. P. et al. P4-ATPases as phospholipid flippases-structure, function, and enigmas. Front. Physiol. 7, 275 (2016)
Montigny, C., Lyons, J., Champeil, P., Nissen, P. & Lenoir, G. On the molecular mechanism of flippase- and scramblase-mediated phospholipid transport. Biochim. Biophys. Acta 1861 (8 Pt B), 767–783 (2016)
Ruiz, N., Kahne, D. & Silhavy, T. J. Transport of lipopolysaccharide across the cell envelope: the long road of discovery. Nat. Rev. Microbiol. 7, 677–683 (2009)
Raetz, C. R., Reynolds, C. M., Trent, M. S. & Bishop, R. E. Lipid A modification systems in gram-negative bacteria. Annu. Rev. Biochem. 76, 295–329 (2007)
Beutler, B. & Rietschel, E. T. Innate immune sensing and its roots: the story of endotoxin. Nat. Rev. Immunol. 3, 169–176 (2003)
Whitfield, C. & Trent, M. S. Biosynthesis and export of bacterial lipopolysaccharides. Annu. Rev. Biochem. 83, 99–128 (2014)
May, J. M., Sherman, D. J., Simpson, B. W., Ruiz, N. & Kahne, D. Lipopolysaccharide transport to the cell surface: periplasmic transport and assembly into the outer membrane. Phil. Trans. R. Soc. Lond. B 370, 20150027 (2015)
Simpson, B. W., May, J. M., Sherman, D. J., Kahne, D. & Ruiz, N. Lipopolysaccharide transport to the cell surface: biosynthesis and extraction from the inner membrane. Phil. Trans. R. Soc. Lond. B 370, 20150029 (2015)
Ward, A., Reyes, C. L., Yu, J., Roth, C. B. & Chang, G. Flexibility in the ABC transporter MsbA: Alternating access with a twist. Proc. Natl Acad. Sci. USA 104, 19005–19010 (2007)
Dong, J., Yang, G. & McHaourab, H. S. Structural basis of energy transduction in the transport cycle of MsbA. Science 308, 1023–1028 (2005)
Zou, P. & McHaourab, H. S. Alternating access of the putative substrate-binding chamber in the ABC transporter MsbA. J. Mol. Biol. 393, 574–585 (2009)
Tarling, E. J., de Aguiar Vallim, T. Q. & Edwards, P. A. Role of ABC transporters in lipid transport and human disease. Trends Endocrinol. Metab. 24, 342–350 (2013)
López-Marqués, R. L. et al. Structure and mechanism of ATP-dependent phospholipid transporters. Biochim. Biophys. Acta 1850, 461–475 (2015)
Locher, K. P. Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat. Struct. Mol. Biol. 23, 487–493 (2016)
Kim, J. et al. Subnanometre-resolution electron cryomicroscopy structure of a heterodimeric ABC exporter. Nature 517, 396–400 (2015)
Oldham, M. L. et al. A mechanism of viral immune evasion revealed by cryo-EM analysis of the TAP transporter. Nature 529, 537–540 (2016)
Zhang, Z. & Chen, J. Atomic structure of the cystic fibrosis transmembrane conductance regulator. Cell 167, 1586–1597.e9 (2016)
Johnson, Z. L. & Chen, J. Structural basis of substrate recognition by the multidrug resistance protein MRP1. Cell 168, 1075–1085.e9 (2017)
Liu, F., Zhang, Z., Csanády, L., Gadsby, D. C. & Chen, J. Molecular structure of the human CFTR ion channel. Cell 169, 85–95.e8 (2017)
Li, N. et al. Structure of a pancreatic ATP-sensitive potassium channel. Cell 168, 101–110.e10 (2017)
Bayburt, T. H. & Sligar, S. G. Membrane protein assembly into nanodiscs. FEBS Lett. 584, 1721–1727 (2010)
Doerrler, W. T. & Raetz, C. R. ATPase activity of the MsbA lipid flippase of Escherichia coli. J. Biol. Chem. 277, 36697–36705 (2002)
Siarheyeva, A. & Sharom, F. J. The ABC transporter MsbA interacts with lipid A and amphipathic drugs at different sites. Biochem. J. 419, 317–328 (2009)
Kawai, T., Caaveiro, J. M., Abe, R., Katagiri, T. & Tsumoto, K. Catalytic activity of MsbA reconstituted in nanodisc particles is modulated by remote interactions with the bilayer. FEBS Lett. 585, 3533–3537 (2011)
Hasin, M. & Kennedy, E. P. Role of phosphatidylethanolamine in the biosynthesis of pyrophosphoethanolamine residues in the lipopolysaccharide of Escherichia coli. J. Biol. Chem. 257, 12475–12477 (1982)
Kaur, H. et al. The ABC exporter MsbA probed by solid state NMR – challenges and opportunities. Biol. Chem. 396, 1135–1149 (2015)
Park, B. S. et al. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature 458, 1191–1195 (2009)
Chen, J. Molecular mechanism of the Escherichia coli maltose transporter. Curr. Opin. Struct. Biol. 23, 492–498 (2013)
Buchaklian, A. H. & Klug, C. S. Characterization of the Walker A motif of MsbA using site-directed spin labeling electron paramagnetic resonance spectroscopy. Biochemistry 44, 5503–5509 (2005)
Zou, P. & McHaourab, H. S. Increased sensitivity and extended range of distance measurements in spin-labeled membrane proteins: Q-band double electron-electron resonance and nanoscale bilayers. Biophys. J. 98, L18–L20 (2010)
Schultz, K. M., Merten, J. A. & Klug, C. S. Characterization of the E506Q and H537A dysfunctional mutants in the E. coli ABC transporter MsbA. Biochemistry 50, 3599–3608 (2011)
Eckford, P. D. & Sharom, F. J. Functional characterization of Escherichia coli MsbA: interaction with nucleotides and substrates. J. Biol. Chem. 283, 12840–12850 (2008)
Cooper, R. S. & Altenberg, G. A. Association/dissociation of the nucleotide-binding domains of the ATP-binding cassette protein MsbA measured during continuous hydrolysis. J. Biol. Chem. 288, 20785–20796 (2013)
Zoghbi, M. E., Cooper, R. S. & Altenberg, G. A. The lipid bilayer modulates the structure and function of an ATP-binding cassette exporter. J. Biol. Chem. 291, 4453–4461 (2016)
Dawson, R. J. & Locher, K. P. Structure of the multidrug ABC transporter Sav1866 from Staphylococcus aureus in complex with AMP-PNP. FEBS Lett. 581, 935–938 (2007)
Choudhury, H. G. et al. Structure of an antibacterial peptide ATP-binding cassette transporter in a novel outward occluded state. Proc. Natl Acad. Sci. USA 111, 9145–9150 (2014)
Lin, D. Y., Huang, S. & Chen, J. Crystal structures of a polypeptide processing and secretion transporter. Nature 523, 425–430 (2015)
Gu, R. X. et al. Conformational changes of the antibacterial peptide ATP binding cassette transporter McjD revealed by molecular dynamics simulations. Biochemistry 54, 5989–5998 (2015)
Doshi, R. et al. Molecular disruption of the power stroke in the ATP-binding cassette transport protein MsbA. J. Biol. Chem. 288, 6801–6813 (2013)
Borbat, P. P. et al. Conformational motion of the ABC transporter MsbA induced by ATP hydrolysis. PLoS Biol. 5, e271 (2007)
Doshi, R. & van Veen, H. W. Substrate binding stabilizes a pre-translocation intermediate in the ATP-binding cassette transport protein MsbA. J. Biol. Chem. 288, 21638–21647 (2013)
Moeller, A. et al. Distinct conformational spectrum of homologous multidrug ABC transporters. Structure 23, 450–460 (2015)
Perez, C. et al. Structure and mechanism of an active lipid-linked oligosaccharide flippase. Nature 524, 433–438 (2015)
Brunner, J. D., Lim, N. K., Schenck, S., Duerst, A. & Dutzler, R. X-ray structure of a calcium-activated TMEM16 lipid scramblase. Nature 516, 207–212 (2014)
Ru, H. et al. Molecular mechanism of V(D)J recombination from synaptic RAG1-RAG2 complex structures. Cell 163, 1138–1152 (2015)
Booth, D. S., Avila-Sakar, A. & Cheng, Y. Visualizing proteins and macromolecular complexes by negative stain EM: from grid preparation to image acquisition. J. Vis. Exp. 58, 3227 (2011)
Li, X., Zheng, S., Agard, D. A. & Cheng, Y. Asynchronous data acquisition and on-the-fly analysis of dose fractionated cryoEM images by UCSFImage. J. Struct. Biol. 192, 174–178 (2015)
Zheng, S., Palovcak, E., Armache, J.-P., Cheng, Y. & Agard, D. Anisotropic correction of beam-induced motion for improved single-particle electron cryo-microscopy. Preprint at http://biorxiv.org/content/early/2016/07/04/061960 (2016)
Mindell, J. A. & Grigorieff, N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 (2003)
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012)
Scheres, S. H. A Bayesian view on cryo-EM structure determination. J. Mol. Biol. 415, 406–418 (2012)
Bai, X. C., Rajendra, E., Yang, G., Shi, Y. & Scheres, S. H. Sampling the conformational space of the catalytic subunit of human γ-secretase. eLife 4, e11182 (2015)
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014)
Lyumkis, D., Brilot, A. F., Theobald, D. L. & Grigorieff, N. Likelihood-based classification of cryo-EM images using FREALIGN. J. Struct. Biol. 183, 377–388 (2013)
Hohl, M., Briand, C., Grütter, M. G. & Seeger, M. A. Crystal structure of a heterodimeric ABC transporter in its inward-facing conformation. Nat. Struct. Mol. Biol. 19, 395–402 (2012)
Guex, N., Peitsch, M. C. & Schwede, T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis 30 (Suppl 1), S162–S173 (2009)
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010)
Adams, P. D . et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010)
van Aalten, D. M. et al. PRODRG, a program for generating molecular topologies and unique molecular descriptors from coordinates of small molecules. J. Comput. Aided Mol. Des. 10, 255–262 (1996)
Kyte, J. & Doolittle, R. F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132 (1982)
Acknowledgements
We thank G. Chang for providing the MsbA expression plasmid. We are grateful to Z. Li and M. Chambers for EM technical support. We thank D. Kahne for making us aware of ClearColi. We thank W. Harper, T. Rapoport and T. Walther for critical reading of the manuscript. We thank members of the Liao group for helpful discussions and comments on the manuscript, and members of the Walz group for help in the initial phases of the project.
Author information
Authors and Affiliations
Contributions
M.L. conceived the project. T.W. advised on experimental design. W.M. purified MsbA, performed nanodisc and proteoliposome reconstitution, generated MsbA mutants, measured ATPase activities, carried out EM data collection and image processing, and built the atomic models. Y.L., S.H.Y. and R.K.E. performed the mass spectrometry measurement and analysis presented in Extended Data Fig. 2g–j. W.M. and M.L. analysed the data and wrote the manuscript with contributions from T.W.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks D. Levy, D. Slotboom and H. W. van Veen for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 LPS and its cross-membrane transport in Gram-negative bacteria.
a, b, Chemical structure (a) and cartoon representation (b) of E. coli rough LPS (lipid A with inner and outer core oligosaccharides). The carbon atoms of the glucosamines are numbered, and the lipid acyl chains attached to the glucosamines are labelled as R2 to R3″. Kdo, 3-deoxy-d-manno-oct-2-ulosonic acid; Hep, l-glycero-d-manno-heptose; Glc, glucose; Gal, galactose. c, The LPS-transport pathway in E. coli consists of four steps: (1) MsbA-driven flipping of rough LPS from the cytoplasmic to the periplasmic leaflet of the inner membrane; (2) WaaL-mediated ligation of O-antigen onto rough LPS to form mature, smooth LPS; (3) cross-periplasm transport of LPS, mediated by proteins LptF, LptG, LptB, LptC and LptA; and (4) LPS insertion into the outer leaflet of the outer membrane, mediated by proteins LptD and LptE.
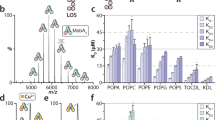
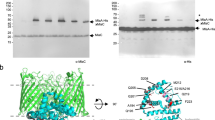
Extended Data Figure 2 Purification and characterization of MsbA in DDM, nanodiscs and proteoliposomes.
a, Gel-filtration profile (Superdex-200) of purified MsbA in nanodiscs formed with MSP1D1 and E. coli polar lipids. Inset shows that the peak material contains His-tagged MsbA (67.2 kDa) and MSP1D1 (22 kDa), and that these migrate as single, homogeneous bands on a Coomassie-stained SDS–PAGE gel. For gel source data, see Supplementary Fig. 1. b, ATPase activities of MsbA in DDM, in nanodiscs formed with different MSPs and lipid compositions and in proteoliposomes. c, Assay of the ATP-concentration-dependent ATPase activity of MsbA in nanodiscs formed with MSP1D1 and E. coli polar lipids yielded a Km of 0.34 ± 0.02 mM and a Vmax of 4.34 ± 0.08 μmol ATP per min per mg MsbA. d, Assay of the ADP-concentration-dependent inhibition of the ATPase activity of MsbA in nanodiscs formed with MSP1D1 and E. coli polar lipids, in the presence of 0.5 mM ATP yielded a Ki(ADP) of 0.54 ± 0.05 mM. e, Assay of the vanadate-concentration-dependent inhibition of the ATPase activity of MsbA in nanodiscs formed with MSP1D1 and E. coli polar lipids, in the presence of 2 mM ATP. With 0.01 mM vanadate, the ATPase activity was inhibited by 90%. f, ATPase activity of wild-type MsbA in proteoliposomes in the absence (−) and presence (+) of 100 μM Kdo2-lipid A. The first two MsbA samples were purified from E. coli strain BL21 (DE3) after membrane solubilization with LDAO or DDM for 1 h, and the third MsbA sample was purified from the ClearColi strain after solubilization with DDM overnight. Each point represents the mean ± s.d. of three separate measurements. The 100% activity is 4.7 ± 0.3 μmol ATP per min per mg MsbA. g, MALDI–TOF mass spectra of buffer alone, Kdo2-lipid A and purified MsbA in DDM. The fragment at m/z 1,796 is consistent with a diphosphorylated hexa-acyl form of lipid A, which is generated by acid hydrolysis to remove the glycosylation on E. coli LPS. h, Tandem MS spectra of the fragments at m/z 1,796 from Kdo2-lipid A and purified MsbA. i, Chemical structures and m/z values of the potential break-down products of E. coli lipid A (m/z = 1,796). j, MALDI–TOF mass spectra of Kdo2-lipid A, wild-type MsbA, and the R78A;R148A;K299A mutant purified in DDM. Samples were prepared as in g.
Extended Data Figure 3 Single-particle EM analysis of the overall architecture of nucleotide-free MsbA in DDM and in nanodiscs formed with different MSPs and lipids.
a, Representative negative-stain EM image and 2D averages of MsbA in DDM. b, Representative negative-stain EM image and 2D averages of MsbA in nanodiscs formed with MSP1D1 and POPG. The box dimension of the 2D averages shown in a and b is 215 Å. c, 2D averages and 3D classification of cryo-EM particles of MsbA reconstituted into nanodiscs with MSP1D1 and E. coli polar lipids. d, 2D averages and 3D classification of cryo-EM particles of MsbA reconstituted into nanodiscs with MSP1D1 and POPG. e, 2D averages and 3D classification of cryo-EM particles of MsbA reconstituted into nanodiscs with MSP1E3D1 and E. coli polar lipids. The box dimension of the 2D averages shown in c–e is 236 Å. Good cryo-EM reconstructions are indicated with blue dotted circles, while other classes yielded distorted 3D reconstructions. f, The superimposition of the 3D reconstructions of class #1 in panel c (grey, indicated by a star) and class #5 in panel e (purple, indicated by a star) shows essentially identical overall architectures of MsbA, despite the very different nanodisc sizes.
Extended Data Figure 4 Image processing workflow for nucleotide-free MsbA in nanodiscs.
a, Three subsequent 3D classifications were used to remove bad particles (classification 1), to select particles with homogeneous TMDs (classification 2), and to exclude particles with very small nanodiscs (classification 3). b, Central cross-sections through the three good cryo-EM reconstructions from the second 3D classification step. Class #3 shows the outer core oligosaccharide. c, Views perpendicular and parallel to the membrane plane of the superimposed cryo-EM reconstructions of class #1 (grey) and class #2 (yellow) from the third 3D classification. Compared to the density map of class #2, the density map of class #1 shows a more open conformation, with its two NBDs moving apart in a shearing motion (black arrows).
Extended Data Figure 5 Single-particle cryo-EM analysis of nucleotide-free MsbA in nanodiscs.
a, 2D averages of cryo-EM particles of MsbA reconstituted into nanodiscs with MSP1D1 and E. coli polar lipids. The particle box dimension is 236 Å. b, Final 3D reconstruction filtered to 4.2 Å (left) and a central cross-section (right), coloured according to the local resolution. c, Gold-standard Fourier shell correlation curves calculated with a soft mask to include only the TMDs (blue curve) or the whole MsbA molecule without the nanodisc (green curve). d, Angular distribution of the cryo-EM particles included in the final reconstruction. e, Selected cryo-EM densities (grey mesh) with the atomic model, showing amino acids that form a ring of hydrophilic interactions surrounding the glucosamines of LPS. Only side chains with well-defined EM density are shown. The main chains are coloured blue and orange to indicate the two MsbA subunits, and LPS is shown in green. The numbers associated with the red dotted lines denote the distances between hydrophilic side chains and the phosphate groups of LPS. f, Protein sequence alignment of TM2, TM3, TM5 and TM6 from Escherichia coli (ECOLI), Salmonella typhimurium (SALTY), Vibrio cholera (VIBCH), Francisella novicida (FRANO), Pseudomonas aeruginosa (PSEAE), Neisseria meningitides (NEIMB), Burkholderia mallei (BURMA) and Bordetella bronchiseptica (BORBR). Only selected regions of the sequences are shown. Important residues with side chains that are represented by clear density in the cryo-EM map are highlighted and labelled according to the residue numbers in E. coli MsbA.
Extended Data Figure 6 Analysis of the cryo-EM structure of nucleotide-free MsbA in nanodiscs.
a, The final cryo-EM reconstruction filtered to 4.2 Å without applying a B-factor with the atomic model of MsbA in the nucleotide-free state. The nanodisc density indicates that the membrane bilayer embeds MsbA in the region between the periplasmic loops and the elbow helices. The green arrow indicates the putative LPS entry site between TM4 and TM6. b, Three perpendicular views of the atomic model of the MsbA TMDs with the cryo-EM density for the LPS molecules. In the top two panels, some helices are omitted for clarity. c, LPS density shown using a normal (left) and a high (right) contouring threshold, together with its atomic model. d, Comparison of the cryo-EM structure of MsbA in nanodiscs (subunits shown in blue and orange) and the crystal structure of E. coli MsbA in detergent (subunits shown in cyan and yellow). The thick blue lines indicate the boundaries of the lipid bilayer for nanodisc-embedded MsbA, and the dashed blue line indicates the level just above the elbow helix in the crystal structure. e, Superimposition of the cryo-EM structure of MsbA in nanodiscs (subunits shown in blue and orange ribbon representation) and the crystal structure of detergent-solubilized Vibrio cholerae MsbA (subunits shown in cyan and yellow wire representation), which was obtained by aligning one TMD/NBD wing from each structure (left panel). Comparison of the non-aligned TMD/NBD wings (right panel) shows that TM4, TM5 and NBD in the crystal structure have moved forward compared to the cryo-EM structure. f, View from the cytoplasm of the cryo-EM structure of MsbA in nanodiscs and the crystal structure of detergent-solubilized Vibrio cholerae MsbA. The numbers associated with the red dotted lines indicate the distances between Cα atoms of Ser378 and Ser482 in the opposing NBDs.
Extended Data Figure 7 Comparison of 3D reconstructions obtained for E. coli MsbA in POPG nanodiscs, MsbA purified from ClearColi in POPG-lipid A nanodiscs and E. coli MsbA in POPC nanodisc.
a, Gold-standard Fourier shell correlation curves (FSC) calculated with a soft mask to include only the TMDs of MsbA. The indicated resolutions were estimated based on the FSC = 0.143 criterion. b, 3D reconstructions filtered to the resolutions of their TMDs, coloured according to the local resolution. c, Cross-sectional views and selected slices through the 3D reconstructions. Below, cartoon representations of E. coli lipid A with inner core, lipid A and lipid IVA are shown. In the slices, the densities representing the phosphorylated glucosamines are indicated by black arrows. The inner core is indicated with red circles in the cross-sectional views and red arrowhead in the slices. Note that the inner core density is completely absent from the cryo-EM map of MsbA purified from ClearColi and reconstituted into nanodiscs with POPG and lipid A. Owing to the low resolution of the acyl chains of lipid A, which is probably caused by their flexibility, it is not clear whether the density inside MsbA purified from ClearColi represents lipid IVA that was possibly co-purified with MsbA or the lipid A that was added during nanodisc reconstitution or a mixture of both.
Extended Data Figure 8 Single-particle cryo-EM analysis of nanodisc-embedded MsbA with ADP-vanadate.
a, Selected area of a representative cryo-EM image. Scale bar indicates 200 Å. b, 2D averages of cryo-EM particles. The particle box dimension is 236 Å. c, Image processing workflow. By applying C2 symmetry, the two best 3D classes were independently refined to 8 and 4.8 Å. The central cross-sections of these two refined EM maps are shown. d, Final 3D reconstruction filtered to 4.8 Å. The map is coloured according to its local resolution. Front view and a central cross-section are shown. e, Gold-standard Fourier shell correlation curve calculated with a soft mask to include only MsbA but not the nanodisc. f, Angular distribution of the cryo-EM particles included in the final 3D reconstruction. g, Comparison of the central cross-sections of the atomic model of nanodisc-reconstituted MsbA with ADP-vanadate and the crystal structure of McjD (PDB ID: 4PL0) in the occluded conformation. TM6 and TM3 are coloured red and magenta, respectively. h, Comparison of the cryo-EM structure of nucleotide-free MsbA in the inward-facing conformation and the crystal structure of MsbA with AMPPNP in the outward-facing conformation. Cross-sections perpendicular to the membrane plane are shown at the level of the LPS glucosamines in nucleotide-free MsbA, with the TM helices numbered and important residues indicated. The green dashed lines indicate the boundaries between the two TMDs. The organization of the TM helices is different in different functional states. Listed below are the distances between the Cα atoms of the indicated pairs of amino acids.
Extended Data Figure 9 Single-particle cryo-EM analysis of nanodisc-embedded MsbA with ADP.
a, Selected area of a representative cryo-EM image. Scale bar indicates 200 Å. b, 2D averages of cryo-EM particles. The particle box dimension is 236 Å. c, Image processing workflow. d, Views of the final 3D reconstruction filtered to 6.9 Å (left) and its central cross-section (right), coloured according to the local resolution. e, Gold-standard Fourier shell correlation curve calculated with a soft mask to include only MsbA but not the nanodisc. f, Angular distribution of the cryo-EM particles included in the final reconstruction. g, Surface view of the cryo-EM reconstruction of nanodisc-embedded MsbA in the presence of ADP (grey), and central cross-sections through the cryo-EM reconstructions of MsbA in the ADP-bound (middle) and nucleotide-free states (right). Both cryo-EM maps were filtered to 6.9 Å resolution and are shown with the atomic model of nanodisc-embedded MsbA in the nucleotide-free state. The green dashed circles indicate LPS densities inside MsbA.
Supplementary information
Supplementary Figure 1
This file contains the uncropped scans with size marker indications (PDF 187 kb)
Proposed mechanism of MsbA-mediated LPS transport
The video shows a morph between the conformations depicted in Figure 5. The MsbA structures in states 2, 5 and 6 are based on the cryo-EM structures of MsbA in the nucleotide-free, vanadate-trapped and ADP-bound states, respectively. The MsbA structures in states 1, 3 and 4 are based on the crystal structures of inward-facing conformation (3B5X), occluded conformation (4PL0), and outward-facing conformation (3B60), respectively. The boundaries of the lipid membrane are indicated with dashed lines. While the movie shows LPS flipping, ATP hydrolysis and the conformational transition of MsbA as separate steps, these changes likely occur as a concerted process. (MP4 7994 kb)
Rights and permissions
About this article
Cite this article
Mi, W., Li, Y., Yoon, S. et al. Structural basis of MsbA-mediated lipopolysaccharide transport. Nature 549, 233–237 (2017). https://doi.org/10.1038/nature23649
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature23649
This article is cited by
-
A new antibiotic traps lipopolysaccharide in its intermembrane transporter
Nature (2024)
-
Structural basis of prostaglandin efflux by MRP4
Nature Structural & Molecular Biology (2024)
-
Probing the allosteric NBD-TMD crosstalk in the ABC transporter MsbA by solid-state NMR
Communications Biology (2024)
-
Dissecting the conformational complexity and mechanism of a bacterial heme transporter
Nature Chemical Biology (2023)
-
Substrate binding-induced conformational transitions in the omega-3 fatty acid transporter MFSD2A
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.