Abstract
The self-association of proteins into symmetric complexes is ubiquitous in all kingdoms of life1,2,3,4,5,6. Symmetric complexes possess unique geometric and functional properties, but their internal symmetry can pose a risk. In sickle-cell disease, the symmetry of haemoglobin exacerbates the effect of a mutation, triggering assembly into harmful fibrils7. Here we examine the universality of this mechanism and its relation to protein structure geometry. We introduced point mutations solely designed to increase surface hydrophobicity among 12 distinct symmetric complexes from Escherichia coli. Notably, all responded by forming supramolecular assemblies in vitro, as well as in vivo upon heterologous expression in Saccharomyces cerevisiae. Remarkably, in four cases, micrometre-long fibrils formed in vivo in response to a single point mutation. Biophysical measurements and electron microscopy revealed that mutants self-assembled in their folded states and so were not amyloid-like. Structural examination of 73 mutants identified supramolecular assembly hot spots predictable by geometry. A subsequent structural analysis of 7,471 symmetric complexes showed that geometric hot spots were buffered chemically by hydrophilic residues, suggesting a mechanism preventing mis-assembly of these regions. Thus, point mutations can frequently trigger folded proteins to self-assemble into higher-order structures. This potential is counterbalanced by negative selection and can be exploited to design nanomaterials in living cells.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Lukatsky, D. B., Shakhnovich, B. E., Mintseris, J. & Shakhnovich, E. I. Structural similarity enhances interaction propensity of proteins. J. Mol. Biol. 365, 1596–1606 (2007)
Levy, E. D., Boeri Erba, E., Robinson, C. V. & Teichmann, S. A. Assembly reflects evolution of protein complexes. Nature 453, 1262–1265 (2008)
André, I., Strauss, C. E., Kaplan, D. B., Bradley, P. & Baker, D. Emergence of symmetry in homooligomeric biological assemblies. Proc. Natl Acad. Sci. USA 105, 16148–16152 (2008)
Schulz, G. E. The dominance of symmetry in the evolution of homo-oligomeric proteins. J. Mol. Biol. 395, 834–843 (2010)
Ahnert, S. E ., Marsh, J. A ., Hernández, H ., Robinson, C. V . & Teichmann, S. A. Principles of assembly reveal a periodic table of protein complexes. Science 350, aaa2245 (2015)
Levy, E. D. & Teichmann, S. Structural, evolutionary, and assembly principles of protein oligomerization. Prog. Mol. Biol. Transl. Sci. 117, 25–51 (2013)
Dykes, G. W., Crepeau, R. H. & Edelstein, S. J. Three-dimensional reconstruction of the 14-filament fibers of hemoglobin S. J. Mol. Biol. 130, 451–472 (1979)
Grueninger, D. et al. Designed protein-protein association. Science 319, 206–209 (2008)
Brodin, J. D., Smith, S. J., Carr, J. R. & Tezcan, F. A. Designed, helical protein nanotubes with variable diameters from a single building block. J. Am. Chem. Soc. 137, 10468–10471 (2015)
Padilla, J. E., Colovos, C. & Yeates, T. O. Nanohedra: using symmetry to design self assembling protein cages, layers, crystals, and filaments. Proc. Natl Acad. Sci. USA 98, 2217–2221 (2001)
Lai, Y. T., Cascio, D. & Yeates, T. O. Structure of a 16-nm cage designed by using protein oligomers. Science 336, 1129 (2012)
King, N. P. et al. Computational design of self-assembling protein nanomaterials with atomic level accuracy. Science 336, 1171–1174 (2012)
Fletcher, J. M. et al. Self-assembling cages from coiled-coil peptide modules. Science 340, 595–599 (2013)
Lanci, C. J. et al. Computational design of a protein crystal. Proc. Natl Acad. Sci. USA 109, 7304–7309 (2012)
Suzuki, Y. et al. Self-assembly of coherently dynamic, auxetic, two-dimensional protein crystals. Nature 533, 369–373 (2016)
Levy, E. D. A simple definition of structural regions in proteins and its use in analyzing interface evolution. J. Mol. Biol. 403, 660–670 (2010)
Eisenberg, D. & Jucker, M. The amyloid state of proteins in human diseases. Cell 148, 1188–1203 (2012)
Knowles, T. P., Vendruscolo, M. & Dobson, C. M. The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell Biol. 15, 384–396 (2014)
Hecht, M. H., Richardson, J. S., Richardson, D. C. & Ogden, R. C. De novo design, expression, and characterization of Felix: a four-helix bundle protein of native-like sequence. Science 249, 884–891 (1990)
DeGrado, W. F., Summa, C. M., Pavone, V., Nastri, F. & Lombardi, A. De novo design and structural characterization of proteins and metalloproteins. Annu. Rev. Biochem. 68, 779–819 (1999)
Doye, J. P., Louis, A. A. & Vendruscolo, M. Inhibition of protein crystallization by evolutionary negative design. Phys. Biol. 1, 9–13 (2004)
Levy, E. D., De, S. & Teichmann, S. A. Cellular crowding imposes global constraints on the chemistry and evolution of proteomes. Proc. Natl Acad. Sci. USA 109, 20461–20466 (2012)
Chothia, C. & Janin, J. Principles of protein–protein recognition. Nature 256, 705–708 (1975)
Jones, S. & Thornton, J. M. Principles of protein-protein interactions. Proc. Natl Acad. Sci. USA 93, 13–20 (1996)
Wodak, S. J. & Janin, J. Structural basis of macromolecular recognition. Adv. Protein Chem. 61, 9–73 (2002)
Kuriyan, J. & Eisenberg, D. The origin of protein interactions and allostery in colocalization. Nature 450, 983–990 (2007)
Narayanaswamy, R. et al. Widespread reorganization of metabolic enzymes into reversible assemblies upon nutrient starvation. Proc. Natl Acad. Sci. USA 106, 10147–10152 (2009)
Li, P. et al. Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340 (2012)
Noree, C., Sato, B. K., Broyer, R. M. & Wilhelm, J. E. Identification of novel filament-forming proteins in Saccharomyces cerevisiae and Drosophila melanogaster. J. Cell Biol. 190, 541–551 (2010)
Gsponer, J. & Babu, M. M. Cellular strategies for regulating functional and nonfunctional protein aggregation. Cell Reports 2, 1425–1437 (2012)
Levy, E. D., Pereira-Leal, J. B., Chothia, C. & Teichmann, S. A. 3D complex: a structural classification of protein complexes. PLoS Comput. Biol. 2, e155 (2006)
Levy, E. D. PiQSi: protein quaternary structure investigation. Structure 15, 1364–1367 (2007)
Mumberg, D., Müller, R. & Funk, M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156, 119–122 (1995)
Klock, H. E., Koesema, E. J., Knuth, M. W. & Lesley, S. A. Combining the polymerase incomplete primer extension method for cloning and mutagenesis with microscreening to accelerate structural genomics efforts. Proteins 71, 982–994 (2008)
Nagai, T. et al. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20, 87–90 (2002)
Knop, M. et al. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15 (10B), 963–972 (1999)
Cohen, Y. & Schuldiner, M. Advanced methods for high-throughput microscopy screening of genetically modified yeast libraries. Methods Mol. Biol. 781, 127–159 (2011)
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012)
Boeker, E. A. & Snell, E. E. Arginine decarboxylase from Escherichia coli. 2. Dissociation and reassociation of subunits. J. Biol. Chem. 243, 1678–1684 (1968)
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005)
Li, X. M. et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods 10, 584–590 (2013)
Mindell, J. A. & Grigorieff, N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 (2003)
Tang, G. et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007)
Scheres, S. H. W. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012)
Jozic, D., Kaiser, J. T., Huber, R., Bode, W. & Maskos, K. X-ray structure of isoaspartyl dipeptidase from E. coli: a dinuclear zinc peptidase evolved from amidohydrolases. J. Mol. Biol. 332, 243–256 (2003)
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63– (2014)
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004)
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012)
Hible, G. et al. Calorimetric and crystallographic analysis of the oligomeric structure of Escherichia coli GMP kinase. J. Mol. Biol. 352, 1044–1059 (2005)
Hindupur, A. et al. The crystal structure of the E. coli stress protein YciF. Protein Sci. 15, 2605–2611 (2006)
Mathews, I. I., Kappock, T. J., Stubbe, J. & Ealick, S. E. Crystal structure of Escherichia coli PurE, an unusual mutase in the purine biosynthetic pathway. Structure 7, 1395–1406 (1999)
Lake, M. W., Temple, C. A., Rajagopalan, K. V. & Schindelin, H. The crystal structure of the Escherichia coli MobA protein provides insight into molybdopterin guanine dinucleotide biosynthesis. J. Biol. Chem. 275, 40211–40217 (2000)
Jaffe, E. K. et al. Species-specific inhibition of porphobilinogen synthase by 4-oxosebacic acid. J. Biol. Chem. 277, 19792–19799 (2002)
Colovos, C., Cascio, D. & Yeates, T. O. The 1.8 Å crystal structure of the ycaC gene product from Escherichia coli reveals an octameric hydrolase of unknown specificity. Structure 6, 1329–1337 (1998)
Thaw, P. et al. Structural insight into gene transcriptional regulation and effector binding by the Lrp/AsnC family. Nucleic Acids Res. 34, 1439–1449 (2006)
Totir, M. et al. Macro-to-micro structural proteomics: native source proteins for high-throughput crystallization. PLoS ONE 7, e32498 (2012)
Auerbach, G. et al. Zinc plays a key role in human and bacterial GTP cyclohydrolase I. Proc. Natl Acad. Sci. USA 97, 13567–13572 (2000)
Thorell, S., Schürmann, M., Sprenger, G. A. & Schneider, G. Crystal structure of decameric fructose-6-phosphate aldolase from Escherichia coli reveals inter-subunit helix swapping as a structural basis for assembly differences in the transaldolase family. J. Mol. Biol. 319, 161–171 (2002)
von Delft, F. et al. Structure of E. coli ketopantoate hydroxymethyl transferase complexed with ketopantoate and Mg2+, solved by locating 160 selenomethionine sites. Structure 11, 985–996 (2003)
Andréll, J. et al. Crystal structure of the acid-induced arginine decarboxylase from Escherichia coli: reversible decamer assembly controls enzyme activity. Biochemistry 48, 3915–3927 (2009)
Lee, K. H. et al. Crystal structures and enzyme mechanisms of a dual fucose mutarotase/ribose pyranase. J. Mol. Biol. 391, 178–191 (2009)
Kanjee, U. et al. Linkage between the bacterial acid stress and stringent responses: the structure of the inducible lysine decarboxylase. EMBO J. 30, 931–944 (2011)
Wang, M. et al. PaxDb, a database of protein abundance averages across all three domains of life. Mol. Cell. Proteomics 11, 492–500 (2012)
Petrovska, I. et al. Filament formation by metabolic enzymes is a specific adaptation to an advanced state of cellular starvation. eLife (2014). Pages?
Landgraf, D., Okumus, B., Chien, P., Baker, T. A. & Paulsson, J. Segregation of molecules at cell division reveals native protein localization. Nat. Methods 9, 480–482 (2012)
DeLano, W. L. The PyMOL molecular graphics system (2002)
Burley, S. K. & Petsko, G. A. Aromatic-aromatic interaction: a mechanism of protein structure stabilization. Science 229, 23–28 (1985)
Singh, J. & Thornton, J. The interaction between phenylalanine rings in proteins. FEBS Lett. 191, 1–6 (1985)
Dougherty, D. A. Cation-pi interactions in chemistry and biology: a new view of benzene, Phe, Tyr, and Trp. Science 271, 163–168 (1996)
Thomas, K. A., Smith, G. M., Thomas, T. B. & Feldmann, R. J. Electronic distributions within protein phenylalanine aromatic rings are reflected by the three-dimensional oxygen atom environments. Proc. Natl Acad. Sci. USA 79, 4843–4847 (1982)
Claverie, P., Hofnung, M. & Monod, J. Sur certaines implications de l’hypothèse d’équivalence stricte entre les protomères des protéines oligomériques. C.R. Acad. Sci. III 266, 1616–1618 (1968)
Lukatsky, D. B., Zeldovich, K. B. & Shakhnovich, E. I. Statistically enhanced self-attraction of random patterns. Phys. Rev. Lett. 97, 178101 (2006)
Acknowledgements
We thank S. Wolf and E. Shimoni for help with electron microscopy experiments, and J. Georgeson for setting up the microscope time-lapse. We thank members of the laboratory, D. Fass and A. Horovitz for discussions throughout the realization of this work, H. Weissman for discussions about electron microscopy, and D. Fass for invaluable feedback on the manuscript. This work was supported by the Israel Science Foundation and the I-CORE Program of the Planning and Budgeting Committee (grants 1775/12 and 2179/14), by the Marie Curie Career Integration Grants Program (number 711715), by the Human Frontier Science Program Career Development Award (number CDA00077/2015), and by a research grant from A.-M. Boucher. H.G.S. received support from the Koshland Foundation and a McDonald-Leapman Grant. Electron microscopy studies were supported by the Irving and Cherna Moskowitz Center for Nano and Bio-Nano Imaging. E.D.L. is incumbent of the Recanati Career Development Chair of Cancer Research.
Author information
Authors and Affiliations
Contributions
H.G.S. and E.D.L. designed the experiments. C.E.M. and E.D.L. designed the computational analyses of protein structure. H.G.S. performed all experiments with guidance from N.E. for electron microscopy techniques. N.E. and H.G.S. performed the single-particle reconstruction. C.E.M. performed the bioinformatics analyses. H.G.S. and E.D.L. analysed the experimental data. C.E.M. and E.D.L. analysed the computational results. E.D.L. and H.G.S. wrote the manuscript with help from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 Increasing surface hydrophobicity of a homo-oligomer triggers its supramolecular self-assembly.
a, Yeast cells expressing a fluorescently tagged homo-octameric dipeptidase from E. coli (PDB accession 1POK). The octamer can be viewed as two rings of four subunits each, stacked tail-to-tail. The localization of the wild-type protein is cytosolic and uniform, but a point mutant (E239Y) triggers head-to-head interactions between octamers and their stacking into micrometre-long fibres. b, Assembly is not mediated by interactions between YFP tags. It has been reported that fluorescent proteins can induce protein aggregation through dimerization65. In our work, none of the wild-type proteins aggregated despite their fusion to YFP, probably because we used the variant bearing the A206K mutation disrupting a weak tendency to dimerize. In addition, we performed a control experiment consisting of co-expressing an excess of the dipeptidase (PDB accession 1POK) untagged with YFP, together with a sub-stoichiometric quantity of the YFP-tagged subunit. As a result, most octamers will harbour zero or one YFP tag. In this context, the YFP allows monitoring the assembly but does not participate in multivalent interactions. c, Fluorescence microscopy revealed that fibres were forming in this context and were about tenfold less fluorescent than the case where all subunits were tagged with YFP. Together, these data indicate that YFP is not mediating fibre assembly in vivo.
Extended Data Figure 2 The proteins studied exhibit intracellular concentrations situated within a physiological range.
a, Intracellular protein concentrations are estimated against reference solutions containing known concentrations of purified YFP. We transferred solutions of purified YFP into the same plate where we inoculated cells for imaging. This enabled us to relate the fluorescence emitted from YFP solutions to that emitted from cells. b, The signal measured by the confocal spinning disk microscope increases linearly with YFP concentration. The equation inferred from linear regression enabled us to convert fluorescence arbitrary units into YFP molarity (1 nM = 0.998 fluorescence arbitrary units − 0.16). c, We used the equation so obtained to transform the median intracellular fluorescence signal into homo-oligomer concentrations. Bars show the population median with associated standard error. We initially used the GPD promoter, which gave concentrations in the sub-micromolar range, and subsequently also used a CYC promoter to express four randomly chosen proteins. Expression with the CYC promoter gave concentrations in the range 10–50 nM, at which mutants also underwent supramolecular assembly (images are shown in Supplementary Table 2).
Extended Data Figure 3 Circular dichroism spectra of all wild-type and mutant pairs studied.
a, We examined the secondary structure content for nine protein pairs using far-ultraviolet circular dichroism. The spectra of wild-type proteins are displayed as black dashed lines and those of mutants as continuous red lines. The measurements were taken at 25 °C. Each pair was compared using the same buffer conditions (Supplementary Table 3). Most of the mutants exhibited similar or identical circular dichroism profiles when compared with their wild-type counterpart, indicating that the content in secondary structure was identical or similar between them. Only the mutants 2WCV (E77Y), 1FRW (D170L/D173L/K175L/D176L) and 1D7A (K11L/E22L/E25L/D158L) showed major differences, but all showed a spectrum with negative ellipticity values in the 210–230 nm range, whereas positive values are expected for random coil. These data indicate that mutants retain a folded structure. b, Stability measurement curves of five wild-type and mutant pairs forming fibres in vivo. Protein stability was assessed for five pairs following the ellipticity at 220 nm from 20 to 85 °C at a heating rate of 1 °C min−1. Wild-type proteins are displayed as black dashed lines and those of mutants as continuous red lines. Each pair was compared using the same buffer conditions (Supplementary Table 3). None of the proteins fully unfolded in the temperature range probed. Thus, we measured the ellipticity of the samples in 2.5 M guanidinium chloride at 90 °C, which was taken as a relative unfolded state (maximal ellipticity, θmax). The ellipticity of the samples at 20 °C was taken as a relative folded state (minimal ellipticity, θmin). We show the normalized ellipticity (θnorm) defined as θnorm = (θT − θmin)/(θmax − θmin), where θT is the ellipticity measured at temperature T.
Extended Data Figure 4 Characterization of protein fibres by electron microscopy.
a, Dipeptidase mutant (1POK E239Y) visualized by TEM with negative staining. The protein buffer was Tris 20 mM, 100 mM NaCl, pH 7.5. The protein mutant self-assembles into filaments that tend to bundle together. b, Example of electron microscopy images on the basis of which the distance separating adjacent homomers in fibres was measured. Mutants in the images are 1D7A (K11L/E22L/E25L/D158L), 2WCV (E77Y), 1L6W (K97Y/K100Y/E102Y), 1M3U (D157L/E158L/D161L), 3N75 (D460L), 2CG4 (K126Y/D131Y), and 1POK (E239Y). All the samples were in Tris 20 mM, pH 7.5 except 1POK (E239Y) which had in addition 100 mM NaCl. c, Fusion of YFP to the dipeptidase mutant (1POK E239Y) does not affect its structure when compared with the wild-type fusion, as seen by circular dichroism. The protein buffer was Tris 20 mM, pH 7.5. d, We examined the dipeptidase fibre-forming mutant fused to YFP by TEM with negative straining. The protein buffer was Tris 20 mM, 100 mM NaCl, pH 7.5. The mutant forms filaments similar to those observed without the YFP fusion.
Extended Data Figure 5 Single-particle three-dimensional reconstruction of the filament formed by the dipeptidase.
a, Structure of the wild-type, octameric enzyme (1POK). b, Structure is rotated along the horizontal axis to show the orientation seen in filaments. c, Representative cryo-electron microscopy image after whole-image motion correction, showing filaments. d, Two-dimensional class averages. e, Final three-dimensional reconstruction. The wild-type structure solved by X-ray crystallography was fitted as a rigid body inside the map, using separate fits for the two tetrameric rings. f, Local resolution map (ResMap) in ångströms. g, Fourier shell correlation for the final reconstruction. The horizontal dotted line indicates the Fourier shell correlation = 0.143 criterion.
Extended Data Figure 6 Detailed view of the helix–helix interface driving the formation of the dipeptidase fibre.
After automated fitting of the atomic coordinates in the electron microscopy density (Extended Data Fig. 5e), we modelled the tyrosine side chain using PYMOL66 and chose the most frequent rotamer. We also translated each helix by 1.15 Å in opposite directions to resolve a steric clash created between the two tyrosines. All other side chains remained unchanged. The resulting model suggests at least three possible types of interaction between side chains, which can help explain how the interface is stabilized. First, a 90° rotation of one tyrosine side chain would enable an aromatic–aromatic interaction, between the negatively charged centre of one tyrosine and the positively charged rim of its neighbour67,68. Second, the arginine of one octamer (R246) could establish a cation–π interaction with the tyrosine of an adjacent octamer69. Finally, the glutamic acid of one octamer (E243) could form an anion–aromatic interaction with the rim of an adjacent tyrosine70.
Extended Data Figure 7 Stickiness is tuned as a function of nDp in dihedral complexes but not in cyclic complexes.
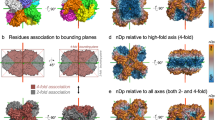
a, The stickiness of an amino acid is defined by the log-ratio of its frequency at protein interfaces relative to protein surfaces. Thus, sticky residues are those enriched at protein interfaces relative to protein surfaces. Stickiness shows strong similarity to hydrophobicity, but also notable differences22. In this analysis, we use stickiness as a measure of ‘interaction propensity’ of surface patches. b, The structure of the dipeptidase (PDB 1POK) is coloured according to nDp calculated with respect to the four-fold axis of symmetry. Structural analyses presented in Fig. 4 consider residues associated with symmetry axes along which fibres can grow, meaning that their nDp values are lowest with respect to those axes. This notion is illustrated with the lower structures, where nDp values of red-coloured residues are smaller with respect to the four-fold axis, while nDp values of residues coloured in grey are smaller with respect to two-fold axes. This strategy enables measuring of negative design along three-, four-, or five-fold axes, while eliminating potential confounding effects due to the two-fold axes. c, Among cyclic complexes, only heterotypic interactions can trigger the formation of infinite fibres71, but such interactions are less likely to form by mutation than by homotypic interactions1,3,4,72. d, We observed that mutations at the ‘tip’ of dihedral complexes are more likely to trigger the formation of supramolecular assemblies than mutations situated farther from the tip, where nDp is larger. Accordingly, we found that stickiness is tuned according to that distance, with regions at greater risk (orange bins, x axis) being associated with lower stickiness (y axis). Lines show the median stickiness of surface patches in any given nDp window and dark-red-coloured error bars correspond to two standard errors. The green dashed line is based on an alternative measure where all residues are counted, irrespective of their distance to two-fold symmetry axes (see Methods). Both measures show that stickiness is tuned as a function of nDp in dihedral homomers. Interestingly, however, we do not observe such tuning in cyclic homomers.
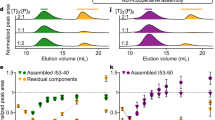
Extended Data Figure 8 Self-assembly takes place at low concentrations in vitro.
a, We assessed whether self-assembly of the dipeptidase mutant takes place at low concentrations. We made serial dilutions of the dipeptidase fused to YFP and then induced self-assembly by addition of 10× PBS to reach a final concentration of 1× PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4). We then incubated the samples for 2 h at 30 °C, spun them down, and pipetted 10 μl of the supernatant to analyse its protein concentration by fluorescence microscopy. b, The concentration of protein in the supernatant relative to the original concentration gave us the fraction of soluble protein. c, The fraction of soluble protein was situated between 0.04 and 0.25 at all concentrations studied, indicating that self-assembly does occur at concentrations as low as 9 nM and probably lower. Error bars span two standard errors and were calculated on the basis of four replicates.
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1 to 4 and Supplementary Text 1. Supplementary Dataset 1 of all homo-oligomeric protein structures with pre-computed nDp measure (>800MB) is available at http://www.weizmann.ac.il/Structural_Biology/faculty_pages/ELevy/downloads/supMat_dPlan.html
Supplementary Table 5
Tabulated information about structural properties of mutated residues
Supplementary Table 6
Tabulated dataset of protein structures and computed descriptors used to generate Fig. 4 and Extended Fig. 7
Cells divide while expressing the fiber-forming mutant of E. coli dipeptidase (1POK E239Y)
Cells divide while expressing the fiber-forming mutant of E. coli dipeptidase. Budding yeasts grow as they express the fiber-forming mutant of E. coli dipeptidase (1pok E239Y) fused to a yellow fluorescent protein. Images were taken every 230 seconds for 10.5 hours. We overlaid the brightfield channel showing the cells (grey) onto the fluorescent channel showing the fibers (green).
Rights and permissions
About this article
Cite this article
Garcia-Seisdedos, H., Empereur-Mot, C., Elad, N. et al. Proteins evolve on the edge of supramolecular self-assembly. Nature 548, 244–247 (2017). https://doi.org/10.1038/nature23320
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature23320
This article is cited by
-
Diverging co-translational protein complex assembly pathways are governed by interface energy distribution
Nature Communications (2024)
-
Mutational biases favor complexity increases in protein interaction networks after gene duplication
Molecular Systems Biology (2024)
-
Emergence of fractal geometries in the evolution of a metabolic enzyme
Nature (2024)
-
Determinants that enable disordered protein assembly into discrete condensed phases
Nature Chemistry (2024)
-
Recording of cellular physiological histories along optically readable self-assembling protein chains
Nature Biotechnology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.