Abstract
The radiation-induced bystander effect (RIBE) refers to a unique process in which factors released by irradiated cells or tissues exert effects on other parts of the animal not exposed to radiation, causing genomic instability, stress responses and altered apoptosis or cell proliferation1,2,3. Although RIBEs have important implications for radioprotection, radiation safety and radiotherapy, the molecular identities of RIBE factors and their mechanisms of action remain poorly understood. Here we use Caenorhabditis elegans as a model in which to study RIBEs, and identify the cysteine protease CPR-4, a homologue of human cathepsin B, as the first RIBE factor in nematodes, to our knowledge. CPR-4 is secreted from animals irradiated with ultraviolet or ionizing gamma rays, and is the major factor in the conditioned medium that leads to the inhibition of cell death and increased embryonic lethality in unirradiated animals. Moreover, CPR-4 causes these effects and stress responses at unexposed sites distal to the irradiated tissue. The activity of CPR-4 is regulated by the p53 homologue CEP-1 in response to radiation, and CPR-4 seems to exert RIBEs by acting through the insulin-like growth factor receptor DAF-2. Our study provides crucial insights into RIBEs, and will facilitate the identification of additional RIBE factors and their mechanisms of action.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Mothersill, C. & Seymour, C. Radiation-induced bystander effects: past history and future directions. Radiat. Res. 155, 759–767 (2001)
Prise, K. M. & O’Sullivan, J. M. Radiation-induced bystander signalling in cancer therapy. Nat. Rev. Cancer 9, 351–360 (2009)
Rzeszowska-Wolny, J., Przybyszewski, W. M. & Widel, M. Ionizing radiation-induced bystander effects, potential targets for modulation of radiotherapy. Eur. J. Pharmacol. 625, 156–164 (2009)
Stergiou, L., Doukoumetzidis, K., Sendoel, A. & Hengartner, M. O. The nucleotide excision repair pathway is required for UV-C-induced apoptosis in Caenorhabditis elegans. Cell Death Differ. 14, 1129–1138 (2007)
Derry, W. B., Putzke, A. P. & Rothman, J. H. Caenorhabditis elegans p53: role in apoptosis, meiosis, and stress resistance. Science 294, 591–595 (2001)
Schumacher, B., Hofmann, K., Boulton, S. & Gartner, A. The C. elegans homolog of the p53 tumor suppressor is required for DNA damage-induced apoptosis. Curr. Biol. 11, 1722–1727 (2001)
Klokov, D. et al. Low dose IR-induced IGF-1-sCLU expression: a p53-repressed expression cascade that interferes with TGFβ1 signaling to confer a pro-survival bystander effect. Oncogene 32, 479–490 (2013)
Koturbash, I. et al. In vivo bystander effect: cranial X-irradiation leads to elevated DNA damage, altered cellular proliferation and apoptosis, and increased p53 levels in shielded spleen. Int. J. Radiat. Oncol. Biol. Phys. 70, 554–562 (2008)
Sun, Y. et al. Treatment-induced damage to the tumor microenvironment promotes prostate cancer therapy resistance through WNT16B. Nat. Med. 18, 1359–1368 (2012)
Buck, M. R., Karustis, D. G., Day, N. A., Honn, K. V. & Sloane, B. F. Degradation of extracellular-matrix proteins by human cathepsin B from normal and tumour tissues. Biochem. J. 282, 273–278 (1992)
Poole, A. R., Tiltman, K. J., Recklies, A. D. & Stoker, T. A. Differences in secretion of the proteinase cathepsin B at the edges of human breast carcinomas and fibroadenomas. Nature 273, 545–547 (1978)
Larminie, C. G. & Johnstone, I. L. Isolation and characterization of four developmentally regulated cathepsin B-like cysteine protease genes from the nematode Caenorhabditis elegans. DNA Cell Biol. 15, 75–82 (1996)
Lorimore, S. A., Rastogi, S., Mukherjee, D., Coates, P. J. & Wright, E. G. The influence of p53 functions on radiation-induced inflammatory bystander-type signaling in murine bone marrow. Radiat. Res. 179, 406–415 (2013)
Calfon, M. et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415, 92–96 (2002)
Bertucci, A., Pocock, R. D., Randers-Pehrson, G. & Brenner, D. J. Microbeam irradiation of the C. elegans nematode. J Radiat. Res. 50, A49–A54 (2009)
Mort, J. S. & Buttle, D. J. Cathepsin B. Int. J. Biochem. Cell Biol. 29, 715–720 (1997)
Shore, D. E. & Ruvkun, G. A cytoprotective perspective on longevity regulation. Trends Cell Biol. 23, 409–420 (2013)
Kenyon, C. J. The genetics of ageing. Nature 464, 504–512 (2010)
Perrin, A. J. et al. Noncanonical control of C. elegans germline apoptosis by the insulin/IGF-1 and Ras/MAPK signaling pathways. Cell Death Differ. 20, 97–107 (2013)
Scott, B. A., Avidan, M. S. & Crowder, C. M. Regulation of hypoxic death in C. elegans by the insulin/IGF receptor homolog DAF-2. Science 296, 2388–2391 (2002)
Paradis, S., Ailion, M., Toker, A., Thomas, J. H. & Ruvkun, G. A PDK1 homolog is necessary and sufficient to transduce AGE-1 PI3 kinase signals that regulate diapause in Caenorhabditis elegans. Genes Dev. 13, 1438–1452 (1999)
Lin, K., Dorman, J. B., Rodan, A. & Kenyon, C. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science 278, 1319–1322 (1997)
Ogg, S. et al. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 389, 994–999 (1997)
Michaelson, D., Korta, D. Z., Capua, Y. & Hubbard, E. J. Insulin signaling promotes germline proliferation in C. elegans. Development 137, 671–680 (2010)
Pinkston, J. M., Garigan, D., Hansen, M. & Kenyon, C. Mutations that increase the life span of C. elegans inhibit tumor growth. Science 313, 971–975 (2006)
Nikjoo, H. & Khvostunov, I. K. A theoretical approach to the role and critical issues associated with bystander effect in risk estimation. Hum. Exp. Toxicol. 23, 81–86 (2004)
Mothersill, C. & Seymour, C. Radiation-induced bystander and other non-targeted effects: novel intervention points in cancer therapy? Curr. Cancer Drug Targets 6, 447–454 (2006)
Recklies, A. D., Tiltman, K. J., Stoker, T. A. & Poole, A. R. Secretion of proteinases from malignant and nonmalignant human breast tissue. Cancer Res. 40, 550–556 (1980)
Barrett, A. J. & Kirschke, H. Cathepsin B, cathepsin H, and cathepsin L. Methods Enzymol. 80, 535–561 (1981)
Shree, T. et al. Macrophages and cathepsin proteases blunt chemotherapeutic response in breast cancer. Genes Dev. 25, 2465–2479 (2011)
Brenner, S. The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974)
Frøkjær-Jensen, C. et al. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 40, 1375–1383 (2008)
Timmons, L., Court, D. L. & Fire, A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263, 103–112 (2001)
Pepper, A. S. R., Killian, D. J. & Hubbard, E. J. A. Genetic analysis of Caenorhabditis elegans glp-1 mutants suggests receptor interaction or competition. Genetics 163, 115–132 (2003)
Bendtsen, J. D., Nielsen, H., von Heijne, G. & Brunak, S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340, 783–795 (2004)
Mello, C. C., Kramer, J. M., Stinchcomb, D. & Ambros, V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10, 3959–3970 (1991)
Gu, T., Orita, S. & Han, M. Caenorhabditis elegans SUR-5, a novel but conserved protein, negatively regulates LET-60 Ras activity during vulval induction. Mol. Cell. Biol. 18, 4556–4564 (1998)
Paquet, C., Sané, A. T., Beauchemin, M. & Bertrand, R. Caspase- and mitochondrial dysfunction-dependent mechanisms of lysosomal leakage and cathepsin B activation in DNA damage-induced apoptosis. Leukemia 19, 784–791 (2005)
Hoogewijs, D., Houthoofd, K., Matthijssens, F., Vandesompele, J. & Vanfleteren, J. R. Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C. elegans. BMC Mol. Biol. 9, 9 (2008)
Acknowledgements
We thank J. Tyler for help with localized irradiation experiments and T. Su for discussion. This work was supported by National Basic Research Program of China (2013CB945602), National Scientific and Technological Major Project of China (2013ZX10002-002), fellowships from China Scholarship Council and Fujian Agriculture and Forestry University (L.Z.) and Tsinghua University-Peking University Center for Life Sciences (Q.L. and X.Z.), and NIH grant R35 GM118188 (D.X.).
Author information
Authors and Affiliations
Contributions
Y.P. set up conditioned medium RIBE assays and performed fractionation, RNAi screen, protein purification, and protease assays. M.Z. and Q.L. generated transgenic animals and conducted experiments with Y.P., aided by H.G. and X.Z. H.L. and L.Z. set up intra-animal RIBE assays and collected related data, aided by Y.-Z.C., X.W. and B.L. J.-T.C. and J.-S.Y. performed mass spectrometry analysis. S.Y. and S.M. generated tm3718. D.X. conceived and supervised the project, analysed data, and wrote the manuscript together with Y.P., M.Z., L.Z., Q.L. and H.L.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks A. Dillin and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 Conditioned medium generated from UV or IR and purified tCPR-4 proteins cause embryonic lethality.
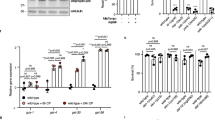
a, The embryonic lethality rate of wild-type (N2) or cep-1(gk138) animals after 100 J m−2 UV irradiation or 500 Gy IR compared with sham-irradiation controls. b, N2 animals were used to generate UV-CM, UV-ctrl, IR-CM and IR-ctrl, which were used to treat unexposed N2 animals in the embryonic lethality assays (Methods). c, Recombinant tCPR-4 (wild type or mutant; 2.8 μM), recombinant human cathepsin B (rhCTSB; 0.27 μM) or the buffer control were used to treat N2 animals in the embryonic lethality assays. Total numbers of embryos scored: 1,781, 805, 1,249, 2,645, 596 and 1,862 embryos, from the left to the right in a; 2,721, 2,484, 880 and 743, from left to right in b; and 979, 875, 929, 939, 907 and 777, from left to right in c. Six independent assays (a, UV-ctrl and UV-CM in b) and three independent assays (IR-ctrl and IR-CM in b, c) were performed for each condition. Data are mean ± s.e.m. NS, not significant; **P < 0.01, ***P < 0.001, two-sided, unpaired t-test.
Extended Data Figure 2 Characterization of the nature and the source of the RIBE factors.
a, b, Treatment of UV-CM and UV-ctrl collected from N2 animals irradiated at 100 J m−2 with RNase (1 μg μl−1) or DNase (0.01 U μl−1) did not alter the apoptosis-inhibitory effect on ced-1(e1735) animals (Methods). Germ-cell corpses were scored after 48-h treatment of ced-1(e1735) L4 larvae. c, e–g, ced-1(e1735) L4 larvae were treated with UV-CM and UV-ctrl (0.1 μg μl−1) prepared from ced-3(n2433) animals (c), glp-1(e2141ts) animals grown at 25 °C (e), N2 animals fed with formaldehyde-treated HB101 bacteria (f), and Pcpr-4::cpr-4::flag; cpr-4(tm3718) animals with or without anti-Flag depletion (g), respectively. Data are mean ± s.e.m. The numbers of gonad arms scored are indicated inside the bars (a–c, e–g). **P < 0.01, ***P < 0.001, two-sided, unpaired t-test. d, Representative differential interference contrast (DIC) images (at least 10) of N2 and glp-1(e2141) adult animals grown at 25 °C. The gonads of the N2 animal with multiple oocytes and fertilized eggs are outlined with dash lines. glp-1(e2141) animal had no visible germ line. Scale bars, 100 μm. h, Immunoblotting analysis of secreted CPR-4::Flag in UV-CM and UV-ctrl prepared from Pcpr-4::cpr-4::flag; cpr-4(tm3718) animals with or without anti-Flag depletion treatment. For gel source data, see Supplementary Fig. 1.
Extended Data Figure 3 Identification of CPR-4 as the RIBE factor.
a, Full medium, the >10 kDa fraction, and the <10 kDa fraction of UV-CM and UV-ctrl derived from N2 animals were used to treat ced-1(e1735) animals in germ-cell corpse assays as in Fig. 1b. Data are mean ± s.e.m. The numbers of gonad arms scored are indicated inside the bars. b, Identification of CPR-4 as the RIBE factor through the RNAi screen. UV-ctrl and UV-CM prepared from RNAi-treated animals were used to treat ced-1(e1735) animals. The number of germ-cell corpse decrease (y-axis) was calculated by subtracting the number of average germ-cell corpses under UV-ctrl treatment from that under UV-CM treatment. Among the candidate genes, RNAi of eft-3, ubq-2 and act-1 caused strong embryonic lethality and we were unable to obtain their UV-CM. RNAi of his-1, his-4 and his-71 caused partial embryonic lethality. 20 gonad arms were scored in each RNAi experiment. c, Secretion of CPR-4::Flag into UV-CM was greatly reduced in irradiated cep-1(gk138) animals carrying a single-copy integration of Pcpr-4::cpr-4::flag compared with that from irradiated N2 animals carrying the same Pcpr-4::cpr-4::flag transgene. Concentrated UV-CM or UV-ctrl (1 μg μl−1) from the indicated strains was subjected to the immunoblotting analysis using an antibody to the Flag epitope. d, The protease activity of 0.27 μM rhCTSB or 2.8 μM recombinant tCPR-4 protein was measured as in Fig. 2b. Data are mean ± s.e.m. (n = 6 in each assay). e, rhCTSB (0.27 μM) or the buffer control was used to treat ced-1(e1735) animals. Animals cultured in the rhCTSB buffer grew slower than in the tCPR-4 buffer and had less germ-cell corpses. Data are mean ± s.e.m. (n = 21 in each assay). Germ-cell corpses were scored after 48 h treatment (a, b, e). ***P < 0.001, two-sided, unpaired t-test. For gel source data, see Supplementary Fig. 1.
Extended Data Figure 4 Representative MS/MS spectra from LTQ-Orbitrap used to confirm the identity of CPR-4 in UV-CM.
a, Tryptic peptides of protein band 6 in the SDS–PAGE gel (Fig. 1d) were analysed by LC–MS/MS using LTQ-Orbitrap. The amino acid sequences of peptides identified by MS/MS analysis and matched to the amino acid sequences of CPR-4 are underlined and in red. b, The MS/MS spectra of the two peptides identified in a are shown. The assignments of the fragmented ions observed to specific amino acid residues were performed using the Scaffold 3 search engine, and the search results are shown below the MS/MS spectra. The lower-case ‘c’ indicates the carbamidomethyl-modified cysteine residue in the tryptic peptide.
Extended Data Figure 5 The cpr-4 deletion mutation and sequence alignment of human and mouse cathepsin B and CPR-4.
a, A schematic representation of the cpr-4 gene structure and the tm3718 deletion. Exons are depicted as blue boxes and introns and the untranslated region as lines. The red box indicates the region of cpr-4 removed by the 406-bp tm3718 deletion. The green box indicates a 12-bp insertion. b, Sequence alignment of human cathepsin B, mouse cathepsin B and CPR-4. Residues that are identical in all three proteins are shaded in pink. The two catalytic residues are shaded in green, which are the active-site cysteine residue that serves as a nucleophile and the histidine residue that acts as a general base to facilitate hydrolysis of the peptide bonds of the substrates16, respectively.
Extended Data Figure 6 Analysis of the roles of additional genes in mediating RIBEs.
a, LUI assays. Animals of the indicated genotype were analysed for the bystander Phsp-4::gfp response 24 h after localized irradiation at the head region as described in Fig. 3. Data are mean ± s.e.m. The numbers of animals scored are indicated inside the bars. b, Germ-cell corpse assays after tCPR-4 treatment. 2.8 μM recombinant tCPR-4 protein or buffer control was used to treat L4 larvae of the indicated genotype as described in Fig. 4a. Data are mean ± s.e.m. The numbers of gonad arms scored are indicated inside the bars. c, Immunoblotting analysis of secreted CPR-4::Flag in UV-CM and UV-ctrl from Pcpr-4::cpr-4::flag; daf-2(e1370); cpr-4(tm3718) animals was done as in Fig. 1f. d, Germ-cell corpse assays. ced-1(e1735) L4 larvae were treated with UV-CM and UV-ctrl (0.1 μg μl−1) prepared from c. Data are mean ± s.e.m. The numbers of gonad arms scored are indicated inside the bars. e, Germ-cell proliferation assays. N2 and cep-1(gk138) L4 larvae were treated in S-medium containing 2.8 μM recombinant tCPR-4 or buffer control for 48 h. The numbers of nuclei and metaphase nuclei in the mitotic zone of the germ line were scored (Methods). Data are mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001, two-sided, unpaired t-test. For gel source data, see Supplementary Fig. 1.
Extended Data Figure 7 The expression patterns of cpr-4 in C. elegans.
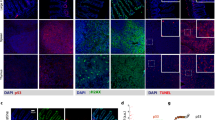
a–g, i, Representative GFP and DIC images (at least 15 each) of N2 animals (a–g) or cep-1(gk138) animals (i) carrying a single-copy integration of Pcpr-4::nls::gfp at the indicated developmental stages. Arrows point to the embryo and the L1 larva that showed no or very dim GFP (a, b). Scale bar, 100 μm. h, Representative DIC, GFP and DIC and GFP merged images (at least 15) of a L4 larva carrying the same Pcpr-4::nls::gfp transgene (left), and corresponding tenfold magnified images showing GFP expression in intestinal cells (right). GFP was seen mostly in the nuclei (arrows). Scale bars, 100 μm (left) and 10 μm (right). j, The intensity of GFP fluorescence in Pcpr-4::nls::gfp and cep-1(gk138); Pcpr-4::nls::gfp animals at different developmental stages was quantified using the Image J software. Data are mean ± s.e.m. n = 28, 28, 24, 31, 30, 33, 52, 52, 19, 28, 52, 52, 24 and 25 animals, scored from left to right, respectively. **P < 0.01, ***P < 0.001, two-sided, unpaired t-test. k, Quantification of GFP intensity in N2 and cep-1(gk138) animals carrying the same single-copy Pcpr-4::nls::gfp transgene irradiated by UV or sham-irradiated, using Image J. Data are mean ± s.e.m. n = 38, 37, 32 and 30 animals, scored from left to right, respectively. ***P < 0.001, two-sided, unpaired t-test.
Extended Data Figure 8 Pharyngeal expression of CPR-4 results in embryonic lethality, larval arrest and reduced germ-cell death.
a, b, Representative DIC and mCherry images (at least 10) of adult animals with pharyngeal expression of CPR-4::mCherry (a) and tCPR-4::mCherry (b). White dashed lines highlight the edge of the pharynx. Arrowheads indicate cells, including coelomocytes, that had taken up CPR-4::mCherry (a), which was made in and secreted from the pharynx and transported to other parts of the animal, probably through the pseudocoelom, a fluid-filled body cavity. The enlarged images of two pairs of posterior cells with weak fluorescence (indicated by colour arrowheads) are shown in dashed boxes with corresponding colours. Scale bars, 100 μm. c, The percentages of embryonic lethality and larval arrest were scored in embryos or larvae carrying Pmyo-2::CPR-4::mCherry (wild-type or mutant) or Pmyo-2::tCPR-4::mCherry transgenes. Three independent transgenic lines were scored for each construct. The number of newly hatched transgenic L1 larvae scored and the number of transgenic embryos scored are indicated in parentheses. The increased larval arrest seen in Pmyo-2::CPR-4::mCherry transgenic animals was blocked when transgenic animals were treated with cpr-4 RNAi (Extended Data Table 2), indicating that reducing cpr-4 expression prevents larval arrest. All animals carry the ced-1(e1735) and cpr-4(tm3718) mutations (a–c). d, Quantification of germ-cell corpses in transgenic animals. L4 ced-1(e1735); cpr-4(tm3718) animals carrying the indicated transgenes were grown on regular NGM plates for 24 h before examination. Data are mean ± s.e.m. The numbers of gonad arms scored are indicated inside the bars. ***P < 0.001, one-way analysis of variance (ANOVA).
Supplementary information
Supplementary Figure
This file contains full scans of the gel images shown in the main and Extended Data Figures.
Source data
Rights and permissions
About this article
Cite this article
Peng, Y., Zhang, M., Zheng, L. et al. Cysteine protease cathepsin B mediates radiation-induced bystander effects. Nature 547, 458–462 (2017). https://doi.org/10.1038/nature23284
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature23284
This article is cited by
-
Investigation into the communication between unheated and heat-stressed Caenorhabditis elegans via volatile stress signals
Scientific Reports (2023)
-
Mechanism and clinical value of exosomes and exosomal contents in regulating solid tumor radiosensitivity
Journal of Translational Medicine (2022)
-
The Role of Bystander Effect in Ultraviolet A Induced Photoaging
Cell Biochemistry and Biophysics (2022)
-
The role of connexin proteins and their channels in radiation-induced atherosclerosis
Cellular and Molecular Life Sciences (2021)
-
Cathepsin B inhibitors block multiple radiation-induced side effects in C. elegans
Cell Research (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.