Abstract
In mammals, chromatin organization undergoes drastic reprogramming after fertilization1. However, the three-dimensional structure of chromatin and its reprogramming in preimplantation development remain poorly understood. Here, by developing a low-input Hi-C (genome-wide chromosome conformation capture) approach, we examined the reprogramming of chromatin organization during early development in mice. We found that oocytes in metaphase II show homogeneous chromatin folding that lacks detectable topologically associating domains (TADs) and chromatin compartments. Strikingly, chromatin shows greatly diminished higher-order structure after fertilization. Unexpectedly, the subsequent establishment of chromatin organization is a prolonged process that extends through preimplantation development, as characterized by slow consolidation of TADs and segregation of chromatin compartments. The two sets of parental chromosomes are spatially separated from each other and display distinct compartmentalization in zygotes. Such allele separation and allelic compartmentalization can be found as late as the 8-cell stage. Finally, we show that chromatin compaction in preimplantation embryos can partially proceed in the absence of zygotic transcription and is a multi-level hierarchical process. Taken together, our data suggest that chromatin may exist in a markedly relaxed state after fertilization, followed by progressive maturation of higher-order chromatin architecture during early development.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Burton, A. & Torres-Padilla, M. E. Chromatin dynamics in the regulation of cell fate allocation during early embryogenesis. Nat. Rev. Mol. Cell Biol. 15, 723–734 (2014)
Gavrilov, A. A. et al. Disclosure of a structural milieu for the proximity ligation reveals the elusive nature of an active chromatin hub. Nucleic Acids Res. 41, 3563–3575 (2013)
Rao, S. S. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014)
Dixon, J. R. et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012)
Battulin, N. et al. Comparison of the three-dimensional organization of sperm and fibroblast genomes using the Hi-C approach. Genome Biol. 16, 77 (2015)
Sasaki, H. & Matsui, Y. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat. Rev. Genet. 9, 129–140 (2008)
Nora, E. P. et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385 (2012)
Lieberman-Aiden, E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009)
Naumova, N. et al. Organization of the mitotic chromosome. Science 342, 948–953 (2013)
Crane, E. et al. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature 523, 240–244 (2015)
Dixon, J. R. et al. Chromatin architecture reorganization during stem cell differentiation. Nature 518, 331–336 (2015)
Wu, J. et al. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature 534, 652–657 (2016)
Qiu, J. J. et al. Delay of ZGA initiation occurred in 2-cell blocked mouse embryos. Cell Res. 13, 179–185 (2003)
Aoki, F., Worrad, D. M. & Schultz, R. M. Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev. Biol. 181, 296–307 (1997)
Schultz, R. M. The molecular foundations of the maternal to zygotic transition in the preimplantation embryo. Hum. Reprod. Update 8, 323–331 (2002)
Abe, K. et al. The first murine zygotic transcription is promiscuous and uncoupled from splicing and 3′ processing. EMBO J. 34, 1523–1537 (2015)
Hug, C. B., Grimaldi, A. G., Kruse, K. & Vaquerizas, J. M. Chromatin architecture emerges during zygotic genome activation independent of transcription. Cell 169, 216–228 (2017)
Nagano, T. et al. Cell cycle dynamics of chromosomal organisation at single-cell resolution. Preprint available at http://biorxiv.org/content/early/2016/12/15/094466 (2016)
Flyamer, I. M. et al. Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature 544, 110–114 (2017)
Brykczynska, U. et al. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat. Struct. Mol. Biol. 17, 679–687 (2010)
Shen, Y. et al. A map of the cis-regulatory sequences in the mouse genome. Nature 488, 116–120 (2012)
Picelli, S. et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protocols 9, 171–181 (2014)
Servant, N. et al. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 16, 259 (2015)
Imakaev, M. et al. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat. Methods 9, 999–1003 (2012)
Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protocols 7, 562–578 (2012)
Akdemir, K. C. & Chin, L. HiCPlotter integrates genomic data with interaction matrices. Genome Biol. 16, 198 (2015)
Zuin, J. et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc. Natl Acad. Sci. USA 111, 996–1001 (2014)
Zhang, B. et al. Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature 537, 553–557 (2016)
Dahl, J. A. et al. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature 537, 548–552 (2016)
Fan, X. et al. Single-cell RNA-seq transcriptome analysis of linear and circular RNAs in mouse preimplantation embryos. Genome Biol. 16, 148 (2015)
Acknowledgements
We thank B. Ren, C. Li, Y. Chen, F. Yue and L. An for discussions, and members of the Xie laboratory for comments. This work was supported by the National Key R&D Program of China (2016YFC0900301 to W.X.), the National Basic Research Program of China (2015CB856201 to W.X.; 2012CB316503 to M.Q.Z.), the National Natural Science Foundation of China (31422031 to W.X.; 61472205 to J.Z.; 31371341 to X.W.; 31361163004 and 31671383 to J.G.; 91519326, 31671384, 31361163004 and 91329000 to M.Q.Z.), the THU-PKU Center for Life Sciences (W.X.), the Youth Thousand Scholar Program of China (W.X., J.Z.), State Key Research Development Program of China (2016YFC1200303 to J.G.), TNLIST Cross-discipline Foundation (M.Q.Z.), the NIH (MH102616 to M.Q.Z.) and Beijing Advanced Innovation Center for Structural Biology (W.X. and J.Z.).
Author information
Authors and Affiliations
Contributions
Z.D. and W.X. conceived the project. Z.D. developed and conducted the sisHi-C experiments. B.H., Z.D., Y.X., and J.M. performed the mouse embryo experiments. Z.D., H.Z., R.M. and J.W. performed bioinformatics analysis. Xi.Z., J.H. and Y.W. helped with the Hi-C analysis pipeline setup. Y.L. and Q.W. performed NGS sequencing. Xu.Z. and K.Z. helped with various experiments. M.Q.Z., J.G., J.R.D., X.W., J.Z. and W.X. supervised experiments or data analyses. Z.D., H.Z. and W.X. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
Z.D. and W.X. are co-inventors on a filed patent application for sisHi-C.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
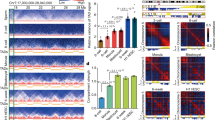
Extended Data Figure 1 Comparison of sisHi-C and in situ Hi-C3.
sisHi-C procedures are shown by the red boxes and arrows on the left, and in situ Hi-C procedures are represented by blue boxes and arrows on the right. The black boxes in the middle show procedures shared by sisHi-C and in situ Hi-C.
Extended Data Figure 2 Validation of sisHi-C data.
a, Hi-C interaction frequency heatmaps (100-kb bin) of chromosome 19 in mouse ES cells as determined by conventional Hi-C4 or sisHi-C using 500 cells (2 replicates). Zoomed-in views (40-kb bin) are also shown. b, Scatter plots showing the comparison of chromatin interaction frequencies between sisHi-C and conventional Hi-C data in mouse ES cells4, or between the two replicates of 500 mouse ES cell sisHi-C samples. Pearson correlation coefficients are also shown. c, Chromatin compartments represented by the first principal component (PC1) values from PCA analyses are shown for the sisHi-C (2 replicates) and conventional Hi-C data in mouse ES cells4. Positive PC1 values represent compartment A regions (yellow), and negative values represent compartment B regions (blue). The gene-dense regions are also shown. d, Bar charts showing the Pearson correlation between the replicates (n = 2–4) of Hi-C data for mouse gametes and early embryos using chromatin interaction frequency. The error bars denote the s.d. of correlation values among different pairs of replicates. e, Heatmaps showing normalized Hi-C interaction frequency (100-kb bin, chromosome 19) in mouse sperm from this study or a previous study5. Zoomed-in views (40-kb bin) are also shown. f, Scatter plots showing the comparison of chromatin interaction frequencies in mouse sperm from this study and a previous study5, with Pearson correlation coefficient shown.
Extended Data Figure 3 Higher-order chromatin structure of MII oocytes.
a, Percentages of inter-chromosomal read pairs among all valid read pairs for gametes and early embryos (replicates pooled, n = 2–4). The error bars denote the s.e.m. among different replicates. b, Hi-C interaction frequency heatmaps comparing the chromatin interaction frequency between human mitotic chromatin (HeLaS3 cells)9 (top right) and mouse MII oocytes (middle right) at 100-kb resolution. The interaction heatmaps for asynchronized human ES cells4 (top left) and mouse ES cells (middle left) are shown as controls. The chromatin contact probabilities relative to genomic distance (P(s) curves) are also shown for each cell type (bottom). P(s) ~ s−0.5 and ~ s−1 represent the predicted mitotic and fractal globule states, respectively. The estimated turning points of chromatin interactions on the P(s) curves for MII oocytes (solid blue) and mitotic HeLaS3 cells (dashed blue) are shown by arrowheads.
Extended Data Figure 4 Establishment of TADs during mouse early development.
a, Hi-C interaction heatmaps (40-kb bin) showing dynamics of local interactions and TADs in mouse preimplantation development (pooled data from 2–4 biological replicates). b, Differential interaction heatmaps showing the changes in interaction frequency between consecutive developmental stages. Regions with increased and decreased interactions from early to late stages (pooled data from 2–4 biological replicates) are shown in yellow and blue, respectively. Hi-C interaction heatmap and identified TADs in ICMs are also shown to indicate the positions of TADs. A pair of subdomains within a TAD that show increased interactions from the 8-cell to ICM stage are indicated by black arrows. c, Overlap of TAD boundaries identified in mouse ES cells (this study) and ICMs (pooled data from three biological replicates). d, Schematic shows the method to compute consolidation score for a TAD. Consolidation score is the ratio between the average intra-TAD interaction frequencies (red) and the average local non-TAD background interaction frequencies in the nearby regions (green). Short-distance interactions (<400 kb) (orange) are excluded. iTF, intra-TAD interaction frequencies; nTF, non-TAD background interaction frequencies. e, Boxplots showing consolidation scores at different developmental stages (pooled data from 2–4 biological replicates). The significance of differences for consolidation scores between two consecutive stages was evaluated by Wilcoxon rank-sum test (two-tailed, with Benjamini–Hochberg multiple testing correction) as shown.
Extended Data Figure 5 TAD boundaries emerge as early as the zygote stage.
a, Hi-C interaction heatmaps showing the chromatin interaction frequencies for TADs in PN5 zygotes (pooled data, n = 4), ICMs (n = 3) and mouse ES cells (n = 2). ICM TADs identified by insulation scores are also shown. Blue boxes show example ICM TAD boundaries. b, The average DI scores of early embryos and mouse ES cells at ICM TAD boundaries (± 500 kb) are shown. DI scores generated by a random valid pair data set are also shown as a control (see Methods).
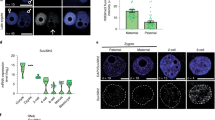
Extended Data Figure 6 Allelic chromatin organization in mouse early development.
a, Boxplots showing the ratios of inter-chromosomal read pairs between maternal and paternal (MP) genomes and between the same parental genomes (MM + PP) (see Methods). The ratios were computed for each pair of different chromosomes (chromosomes 1–2, 1–3 and so on) separately. Boxplots show the variations across all such pairs of chromosomes. b, Percentages of distal intra-chromosomal read pairs (>2 Mb) assigned to the paternal or maternal genome. The significance of allelic imbalance was assessed by comparison with that of the cortex (control) using a two-tailed t-test; **P < 0.001; *P < 0.05; ns, not significant. c, Heatmaps showing the allelic chromatin interaction frequencies (300-kb bin, chromosome 15) of sperm (replicates pooled, n = 3), MII oocyte (n = 2), PN3 (n = 2) and PN5 (n = 2) zygotes. d, Heatmaps showing the normalized average interaction frequencies for all ICM TADs as well as their nearby regions for sperm, MII oocyte and each allele of PN3 zygotes, PN5 zygotes and ICMs.
Extended Data Figure 7 Establishment of chromatin compartments during mouse early development.
a, Pearson correlation coefficients of correlation matrices between each developmental stage (replicates pooled, n = 2–4) and ICM (n = 3) on each allele. b, Hierarchical clustering based on correlation matrix for the two parental alleles. c, UCSC genome browser view showing the enrichment of ATAC-seq and chromatin compartments (PC1 values) for two parental alleles. Gene density is also shown. d, Allelic expression levels of ZGA-only genes (FPKM <0.5 in oocyte and FPKM >1 at any preimplantation stage) that fall into either compartment A or B in early development. The significance of differences between compartments A and B for each allele of each stage was assessed using Wilcoxon rank-sum test (P < 0.02 in all cases, two-tailed, with Benjamini–Hochberg multiple testing correction). e, ATAC-seq enrichment for compartments A and B of two parental alleles in early development. The significance of differences between compartments A and B for each allele of each stage was assessed using two-tailed t-test (P < 5.4 × 10−45 in all cases, with Benjamini–Hochberg multiple testing correction).
Extended Data Figure 8 Chromatin organization establishment in early development can partially proceed in the absence of zygotic genome activation.
a, Gene expression levels (log2[FPKM+1]) show the effects of alpha-amanitin on genes activated during ZGA, genes expressed at all stages or genes repressed at all stages as defined previously28. P values calculated with two-tailed Wilcoxon rank-sum test are shown. Note that genes expressed at all stages presumably have both oocyte-inherited transcripts and zygotically transcribed transcripts. Their decreasing values from 20 h to 45 h in the presence of alpha-amanitin are consistent with continual maternal RNA degradation. Previous RNA-seq data sets29,30 were similarly analysed as controls. b, Heatmaps showing the chromatin interaction frequency (300-kb bin, chromosome 13) for embryos treated with or without alpha-amanitin (replicates pooled, n = 2) (see Methods). The PN3 zygotes were treated with alpha-amanitin and collected for Hi-C analyses at 20 h or 45 h. In both cases the embryos were arrested at the late 2-cell stage. The control groups developed to the late 2-cell and 8-cell stage, respectively. c, Heatmaps showing the normalized average interaction frequencies for all ICM TADs as well as their nearby regions (± 0.5 TAD length) for embryos treated with or without alpha-amanitin (replicates pooled, n = 2) (bottom). Differential interaction heatmap between embryos treated for 20 h and 45 h (top) is also shown. d, Boxplots showing the consolidation scores for embryos treated with alpha-amanitin for 20 h or 45 h for each of the two replicates. P values calculated with two-tailed Wilcoxon rank-sum test are shown.
Extended Data Figure 9 Hierarchical packaging of chromatin in early development.
a, Heatmaps showing the intra-chromosomal interaction probability relative to genomic distance between interacting regions (P(s)) across the whole genome. The P(s) ~ s−1 values are also shown as a reference. b, Heatmaps showing normalized P(s) distributions generated by normalizing P(s) values for each stage to the P(s) ~ s−1 values. c, Hi-C interaction heatmaps showing normalized Hi-C interaction frequency (100-kb bin, chromosome 5). Dashed boxes indicate various regions where chromatin compaction occurs.
Extended Data Figure 10 Hierarchical packaging of chromatin in early development.
a, The contact probability as a function of genomic distance (P(s) analysis) (averaged across all chromosomes) is shown for gametes and zygotes on each allele as previously described9. P(s) ~ s−0.5 and P(s) ~ s−1 are shown to represent the predicted mitotic and fractal globule states, respectively9. b–e, P(s) curves (averaged across all chromosomes) for every two consecutive developmental stages calculated for each allele. P(s) ~ s−1 is shown to represent the predicted fractal globule state9. Arrowheads indicate estimated turning points on the P(s) curves where chromatin interactions drop abruptly (interaction insulation boundary). f, P(s) analyses for individual chromosomes (1, 4, 14 and 19) at early 2-cell stage. P(s) ~ s−1 is shown to represent the predicted fractal globule state. The arrowhead indicates the interaction insulation boundary.
Supplementary information
Supplementary Table
This table contains details of the sequencing information of sisHi-C libraries.
Rights and permissions
About this article
Cite this article
Du, Z., Zheng, H., Huang, B. et al. Allelic reprogramming of 3D chromatin architecture during early mammalian development. Nature 547, 232–235 (2017). https://doi.org/10.1038/nature23263
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature23263
This article is cited by
-
The impact of DNA methylation on CTCF-mediated 3D genome organization
Nature Structural & Molecular Biology (2024)
-
Cell-type differential targeting of SETDB1 prevents aberrant CTCF binding, chromatin looping, and cis-regulatory interactions
Nature Communications (2024)
-
Emergence of replication timing during early mammalian development
Nature (2024)
-
RAD21 is the core subunit of the cohesin complex involved in directing genome organization
Genome Biology (2023)
-
Pan-3D genome analysis reveals structural and functional differentiation of soybean genomes
Genome Biology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.