Abstract
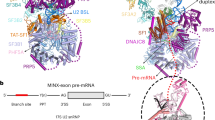
Intron removal requires assembly of the spliceosome on precursor mRNA (pre-mRNA) and extensive remodelling to form the spliceosome’s catalytic centre. Here we report the cryo-electron microscopy structure of the yeast Saccharomyces cerevisiae pre-catalytic B complex spliceosome at near-atomic resolution. The mobile U2 small nuclear ribonucleoprotein particle (snRNP) associates with U4/U6.U5 tri-snRNP through the U2/U6 helix II and an interface between U4/U6 di-snRNP and the U2 snRNP SF3b-containing domain, which also transiently contacts the helicase Brr2. The 3′ region of the U2 snRNP is flexibly attached to the SF3b-containing domain and protrudes over the concave surface of tri-snRNP, where the U1 snRNP may reside before its release from the pre-mRNA 5′ splice site. The U6 ACAGAGA sequence forms a hairpin that weakly tethers the 5′ splice site. The B complex proteins Prp38, Snu23 and Spp381 bind the Prp8 N-terminal domain and stabilize U6 ACAGAGA stem–pre-mRNA and Brr2–U4 small nuclear RNA interactions. These results provide important insights into the events leading to active site formation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Will, C. L. & Lührmann, R. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 3, a003707 (2011)
Boesler, C. et al. A spliceosome intermediate with loosely associated tri-snRNP accumulates in the absence of Prp28 ATPase activity. Nat. Commun. 7, 11997 (2016)
Staley, J. P. & Guthrie, C. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol. Cell 3, 55–64 (1999)
Lesser, C. F. & Guthrie, C. Mutations in U6 snRNA that alter splice site specificity: implications for the active site. Science 262, 1982–1988 (1993)
Lybarger, S. et al. Elevated levels of a U4/U6.U5 snRNP-associated protein, Spp381p, rescue a mutant defective in spliceosome maturation. Mol. Cell. Biol. 19, 577–584 (1999)
Stevens, S. W. & Abelson, J. Purification of the yeast U4/U6.U5 small nuclear ribonucleoprotein particle and identification of its proteins. Proc. Natl Acad. Sci. USA 96, 7226–7231 (1999)
Laggerbauer, B., Achsel, T. & Lührmann, R. The human U5-200kD DEXH-box protein unwinds U4/U6 RNA duplices in vitro. Proc. Natl Acad. Sci. USA 95, 4188–4192 (1998)
Raghunathan, P. L. & Guthrie, C. RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr. Biol. 8, 847–855 (1998)
Fabrizio, P. et al. The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol. Cell 36, 593–608 (2009)
Bessonov, S. et al. Characterization of purified human Bact spliceosomal complexes reveals compositional and morphological changes during spliceosome activation and first step catalysis. RNA 16, 2384–2403 (2010)
Tarn, W. Y. et al. Functional association of essential splicing factor(s) with PRP19 in a protein complex. EMBO J. 13, 2421–2431 (1994)
Hoskins, A. A., Rodgers, M. L., Friedman, L. J., Gelles, J. & Moore, M. J. Single molecule analysis reveals reversible and irreversible steps during spliceosome activation. eLife 5, e14166 (2016)
Madhani, H. D. & Guthrie, C. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell 71, 803–817 (1992)
Steitz, T. A. & Steitz, J. A. A general two-metal-ion mechanism for catalytic RNA. Proc. Natl Acad. Sci. USA 90, 6498–6502 (1993)
Fica, S. M. et al. RNA catalyses nuclear pre-mRNA splicing. Nature 503, 229–234 (2013)
Newman, A. J. & Norman, C. U5 snRNA interacts with exon sequences at 5′ and 3′ splice sites. Cell 68, 743–754 (1992)
Sontheimer, E. J. & Steitz, J. A. The U5 and U6 small nuclear RNAs as active site components of the spliceosome. Science 262, 1989–1996 (1993)
Galej, W. P. et al. Cryo-EM structure of the spliceosome immediately after branching. Nature 537, 197–201 (2016)
Wan, R., Yan, C., Bai, R., Huang, G. & Shi, Y. Structure of a yeast catalytic step I spliceosome at 3.4 Å resolution. Science 353, 895–904 (2016)
Rauhut, R. et al. Molecular architecture of the Saccharomyces cerevisiae activated spliceosome. Science 353, 1399–1405 (2016)
Yan, C., Wan, R., Bai, R., Huang, G. & Shi, Y. Structure of a yeast activated spliceosome at 3.5 Å resolution. Science 353, 904–911 (2016)
Yan, C., Wan, R., Bai, R., Huang, G. & Shi, Y. Structure of a yeast step II catalytically activated spliceosome. Science 355, 149–155 (2017)
Fica, S. M. et al. Structure of a spliceosome remodelled for exon ligation. Nature 542, 377–380 (2017)
Bertram, K. et al. Cryo-EM structure of a human spliceosome activated for step 2 of splicing. Nature 542, 318–323 (2017)
Agafonov, D. E. et al. Molecular architecture of the human U4/U6.U5 tri-snRNP. Science 351, 1416–1420 (2016)
Wan, R. et al. The 3.8 Å structure of the U4/U6.U5 tri-snRNP: insights into spliceosome assembly and catalysis. Science 351, 466–475 (2016)
Nguyen, T. H. D. et al. Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 Å resolution. Nature 530, 298–302 (2016)
van Roon, A.-M. M. et al. Crystal structure of U2 snRNP SF3b components: Hsh49p in complex with Cus1p-binding domain. RNA 23, 968–981 (2017)
Lin, P.-C. & Xu, R.-M. Structure and assembly of the SF3a splicing factor complex of U2 snRNP. EMBO J. 31, 1579–1590 (2012)
Cretu, C. et al. Molecular architecture of SF3b and structural consequences of its cancer-related mutations. Mol. Cell 64, 307–319 (2016)
Rigo, N., Sun, C., Fabrizio, P., Kastner, B. & Lührmann, R. Protein localisation by electron microscopy reveals the architecture of the yeast spliceosomal B complex. EMBO J. 34, 3059–3073 (2015)
Boehringer, D. et al. Three-dimensional structure of a pre-catalytic human spliceosomal complex B. Nat. Struct. Mol. Biol. 11, 463–468 (2004)
Deckert, J. et al. Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol. Cell. Biol. 26, 5528–5543 (2006)
Tarn, W.-Y., Lee, K.-R. & Cheng, S. C. Yeast precursor mRNA processing protein PRP19 associates with the spliceosome concomitant with or just after dissociation of U4 small nuclear RNA. Proc. Natl Acad. Sci. USA 90, 10821–10825 (1993)
Wolf, E. et al. Exon, intron and splice site locations in the spliceosomal B complex. EMBO J. 28, 2283–2292 (2009)
Krämer, A., Grüter, P., Gröning, K. & Kastner, B. Combined biochemical and electron microscopic analyses reveal the architecture of the mammalian U2 snRNP. J. Cell Biol. 145, 1355–1368 (1999)
Nesic, D. & Krämer, A. Domains in human splicing factors SF3a60 and SF3a66 required for binding to SF3a120, assembly of the 17S U2 snRNP, and prespliceosome formation. Mol. Cell. Biol. 21, 6406–6417 (2001)
Igel, H., Wells, S., Perriman, R. & Ares, M. Jr. Conservation of structure and subunit interactions in yeast homologues of splicing factor 3b (SF3b) subunits. RNA 4, 1–10 (1998)
Pauling, M. H., McPheeters, D. S. & Ares, M. Jr. Functional Cus1p is found with Hsh155p in a multiprotein splicing factor associated with U2 snRNA. Mol. Cell. Biol. 20, 2176–2185 (2000)
Dybkov, O. et al. U2 snRNA-protein contacts in purified human 17S U2 snRNPs and in spliceosomal A and B complexes. Mol. Cell. Biol. 26, 2803–2816 (2006)
Schneider, C. et al. Dynamic contacts of U2, RES, Cwc25, Prp8 and Prp45 proteins with the pre-mRNA branch-site and 3′ splice site during catalytic activation and step 1 catalysis in yeast spliceosomes. PLoS Genet. 11, e1005539 (2015)
Xie, J., Beickman, K., Otte, E. & Rymond, B. C. Progression through the spliceosome cycle requires Prp38p function for U4/U6 snRNA dissociation. EMBO J. 17, 2938–2946 (1998)
Stevens, S. W. et al. Biochemical and genetic analyses of the U5, U6, and U4/U6 x U5 small nuclear ribonucleoproteins from Saccharomyces cerevisiae. RNA 7, 1543–1553 (2001)
Agafonov, D. E. et al. Semiquantitative proteomic analysis of the human spliceosome via a novel two-dimensional gel electrophoresis method. Mol. Cell. Biol. 31, 2667–2682 (2011)
Gottschalk, A. et al. Identification by mass spectrometry and functional analysis of novel proteins of the yeast [U4/U6·U5] tri-snRNP. EMBO J. 18, 4535–4548 (1999)
Ulrich, A. K. C., Seeger, M., Schütze, T., Bartlick, N. & Wahl, M. C. Scaffolding in the spliceosome via single α helices. Structure 24, 1972–1983 (2016)
Kuhn, A. N., Li, Z. & Brow, D. A. Splicing factor Prp8 governs U4/U6 RNA unwinding during activation of the spliceosome. Mol. Cell 3, 65–75 (1999)
Chan, S.-P. & Cheng, S.-C. The Prp19-associated complex is required for specifying interactions of U5 and U6 with pre-mRNA during spliceosome activation. J. Biol. Chem. 280, 31190–31199 (2005)
Fica, S. M., Mefford, M. A., Piccirilli, J. A. & Staley, J. P. Evidence for a group II intron-like catalytic triplex in the spliceosome. Nat. Struct. Mol. Biol. 21, 464–471 (2014)
Shuster, E. O. & Guthrie, C. Two conserved domains of yeast U2 snRNA are separated by 945 nonessential nucleotides. Cell 55, 41–48 (1988)
Nguyen, T. H. D. et al. The architecture of the spliceosomal U4/U6.U5 tri-snRNP. Nature 523, 47–52 (2015)
Umen, J. G. & Guthrie, C. A novel role for a U5 snRNP protein in 3′ splice site selection. Genes Dev. 9, 855–868 (1995)
Abelson, J., Hadjivassiliou, H. & Guthrie, C. Preparation of fluorescent pre-mRNA substrates for an smFRET study of pre-mRNA splicing in yeast. Methods Enzymol. 472, 31–40 (2010)
Wu, T. P., Ruan, K. C. & Liu, W. Y. A fluorescence-labeling method for sequencing small RNA on polyacrylamide gel. Nucleic Acids Res. 24, 3472–3473 (1996)
Zhou, Z. & Reed, R. Purification of functional RNA-protein complexes using MS2-MBP. Curr. Protoc. Mol. Biol. 63, 27.3.1—27.3.7 (2001)
Li, X. et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods 10, 584–590 (2013)
Rohou, A. & Grigorieff, N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015)
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012)
Scheres, S. H. W. & Chen, S. Prevention of overfitting in cryo-EM structure determination. Nat. Methods 9, 853–854 (2012)
Tang, G. et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007)
Scheres, S. H. Semi-automated selection of cryo-EM particles in RELION-1.3. J. Struct. Biol. 189, 114–122 (2015)
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004)
Bai, X.-C., Rajendra, E., Yang, G., Shi, Y. & Scheres, S. H. W. Sampling the conformational space of the catalytic subunit of human γ-secretase. eLife 4, e11182 (2015)
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Adams, P. D . et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Zhou, L. et al. Crystal structures of the Lsm complex bound to the 3′ end sequence of U6 small nuclear RNA. Nature 506, 116–120 (2014)
Leung, A. K. W., Nagai, K. & Li, J. Structure of the spliceosomal U4 snRNP core domain and its implication for snRNP biogenesis. Nature 473, 536–539 (2011)
Eswar, N . et al. Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinformatics Chapter 5, 5.6.1–5.6.3 (2014)
Wriggers, W. Conventions and workflows for using Situs. Acta Crystallogr. D 68, 344–351 (2012)
Lorenz, R. et al. ViennaRNA Package 2.0. Algorithms Mol. Biol. 6, 26 (2011)
Popenda, M. et al. Automated 3D structure composition for large RNAs. Nucleic Acids Res. 40, e112 (2012)
Mozaffari-Jovin, S. et al. Inhibition of RNA helicase Brr2 by the C-terminal tail of the spliceosomal protein Prp8. Science 341, 80–84 (2013)
McGrail, J. C. & O’Keefe, R. T. The U1, U2 and U5 snRNAs crosslink to the 5′ exon during yeast pre-mRNA splicing. Nucleic Acids Res. 36, 814–825 (2008)
Sharma, S., Wongpalee, S. P., Vashisht, A., Wohlschlegel, J. A. & Black, D. L. Stem-loop 4 of U1 snRNA is essential for splicing and interacts with the U2 snRNP-specific SF3A1 protein during spliceosome assembly. Genes Dev. 28, 2518–2531 (2014)
van Nues, R. W. & Beggs, J. D. Functional contacts with a range of splicing proteins suggest a central role for Brr2p in the dynamic control of the order of events in spliceosomes of Saccharomyces cerevisiae. Genetics 157, 1451–1467 (2001)
Acknowledgements
We thank C. Savva, S. Chen, G. Cannone, K. R. Vinothkumar, G. McMullan, J. Grimmett and T. Darling for maintaining electron microscopy and computing facilities; the mass spectrometry facility for protein identification, and A. Newman, L. Strittmatter, S. Fica, M. Wilkinson and C. Norman for help and critical reading of the manuscript. We thank J. Löwe, V. Ramakrishnan, D. Barford and R. Henderson for their continuing support. The project was supported by the Medical Research Council (MC_U105184330) and European Research Council Advanced Grant (693087 - SPLICE3D). C.P. was supported by an EMBO Long-Term Fellowship.
Author information
Authors and Affiliations
Contributions
C.P. established complex preparation. C.P. and P.-C.L. performed cryo-EM structure determination and model building. C.P. refined the model. C.P., P.-C.L. and K.N. analysed the structure and wrote the manuscript. K.N. initiated and supervised the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 Biochemical characterization and cryo-EM of the B complex spliceosome.
a, Protein analysis of purified B complex (SDS–PAGE stained with Coomassie blue). U1 snRNP components and U1 snRNA are sub-stoichiometric (see b), consistent with U1 snRNP destabilization in B complex2,31. For gel source data see Supplementary Fig. 1a. b, RNA analysis of purified B complex (denaturing 9% polyacrylamide TBE gel stained with Toluidine blue). For gel source data see Supplementary Fig. 1b. c, Purified B complex is active in an in vitro splicing assay. Splicing reactions were carried out in yeast extract in the absence (lanes 1 and 2) or presence of 60 nM unlabelled competitor pre-mRNA (lane 3), prohibiting the assembly of new spliceosomes. B complex was assembled on labelled pre-mRNA and purified (see Methods), and added to yeast extract for 10 min together with 60 nM unlabelled competitor pre-mRNA, before the addition of ATP to initiate the reaction (lanes 4 and 5). Splicing reactions contained 2 mM ATP (lanes 1–5). The asterisk marks a degradation product. For gel source data see Supplementary Fig. 1c. d, Cryo-EM micrograph of B complex. Scale bar, 200 nm. e, Representative 2D class averages of B complex reveal flexibility of peripheral regions relative to the tri-snRNP body. f, Composite cryo-EM density of B complex shown in two orthogonal views. Colours indicate the respective cryo-EM densities used for modelling (B1, green; B2, yellow; B3, light green; B4, grey; B5, blue; B6, light blue; B7, magenta). The sharpened densities are shown and are aligned using overlapping regions (see Extended Data Fig. 2). The percentage of particles from the full set of 496,581 that contribute to the respective density is indicated together with the overall resolution (see Extended Data Fig. 9). g, Composite cryo-EM density of B complex superimposed on a ribbon model of the B complex structure, coloured as in Fig. 1. The B complex, excluding the U1 snRNP, has a molecular mass of 2.5 MDa of which we modelled 1.8 MDa.
Extended Data Figure 2 Three-dimensional classification of cryo-EM data.
Three-dimensional image classification of the cryo-EM dataset using the B complex negative-stain reconstruction (Methods; Extended Data Fig. 3b) as the initial reference model. The percentage of single particles contributing to each class is provided. To help visualize structural differences, 3D reconstructions of B complex are coloured according to mobile regions: SF3b (green), U2 3′ domain–SF3a (light green), helicase (cyan), body (grey), foot (navy blue), and B complex proteins (magenta). For each classification round, the type of mask and use of signal subtraction is indicated. Additionally, the type of mask, overall resolution, and use of signal subtraction is also indicated for each 3D refinement after classification. For additional details, see the Methods and Extended Data Fig. 9.
Extended Data Figure 3 Negative-stain and cryo-EM reconstructions of B complex.
a, Three-dimensional image classification of negative-stain EM data. A total of 12,296 particles were refined using the negative-stain reconstruction of the human BΔU1-complex32 (EMDB code EMD-1066) as the initial reference, and were subsequently classified. Class 2 contained most features and was used for 3D refinement. The percentage of single particles contributing to each class is provided. b, Two orthogonal views of the yeast B complex negative-stain reconstruction used as the initial reference for processing of the cryo-EM dataset. c, Gold-standard Fourier shell correlation (FSC = 0.143) of the respective B1, B2, B3, B4, B5, B6 and B7 cryo-EM single-particle reconstructions. d, Orientation distribution plot of all particles that contribute to the respective B1, B2, B3, B4, B5, B6 and B7 cryo-EM single-particle reconstructions. e, The composite B complex cryo-EM density (maps B1–B7) is shown in two orthogonal views and coloured by local resolution, as determined by ResMap64. Compare with Extended Data Fig. 1f. f, A central slice through the composite B complex cryo-EM density (maps B1–B7) is shown in two orthogonal views and coloured by local resolution, as determined by ResMap64.
Extended Data Figure 4 Details of the U2 snRNP.
a, Multiple conformations of the U2 snRNP 3′ domain–SF3a subcomplexes relative to SF3b indicate flexibility. This apparent mobility may be important for formation of the A complex, consistent with dynamic contacts of the U2 snRNA 5′ end with SF3a60 (human PRP9) and SF3b49 (human HSH49) in the isolated U2 snRNP40 that differ from U2 snRNP protein–snRNA interactions observed in the yeast B complex structure. Surface representations of the U2 3′ domain (light green), SF3a (light yellow), and SF3b (dark green) are shown. The U2 3′ domain–SF3a complex is positioned according to 3D classifications 1, 6 and 7 from round 5 (compare with Extended Data Fig. 2). A cartoon summarizes the movements. b, Representative regions of the sharpened SF3b-containing density (B1) at 3.9 Å resolution are superimposed on the refined coordinate model. The density shows side-chain features for a loop in Cus1, a β-strand in Rse1, an α-helix in Hsh155, and separation of RNA nucleotides in the U2–pre-mRNA branch helix. Colours are as in Fig. 2. c, Cryo-EM densities for SF3a are superimposed on the B complex coordinate model. The crystal structure of the Y-shaped core of SF3a (ref. 29) is superimposed on the B3 density. Prp11 ZnF, Prp9 ZnF2 and the Prp9 C terminus are superimposed on the B1 density. Structural elements of SF3a, including the Prp9 wedge helix, and disordered regions are indicated. For cryo-EM density nomenclature, see Extended Data Fig. 1f. Colours are as in Fig. 2.
Extended Data Figure 5 Flexibility of the U2 snRNP relative to tri-snRNP.
a, Multiple positions of the U2 snRNP relative to tri-snRNP. Representative classes are shown (classes 1, 6 and 8 from round 3, see Extended Data Fig. 2) that reside along a continuum of conformations (compare with Extended Data Fig. 1e). The U2 snRNP moves together with the U6 LSm ring, which is anchored via the putative Prp3 N terminus. A cartoon summarizes the movements in two orthogonal views. The location of the U2 snRNP has an apparent effect on the strength of the putative pre-mRNA 5′ exon cryo-EM density (compare Extended Data Fig. 6e). When the U2 snRNP is positioned away from the U6 ACAGAGA stem, the pre-mRNA density is weaker than when the U2 snRNP is positioned closer. This suggests how Brr2 helicase activity may perform a kinetic proofreading of the pre-mRNA 5′-exon–U6 snRNA interaction: When the U2 snRNP is close to the tri-snRNP, activation may occur normally (productive activation). However, when the U2 snRNP is positioned further away and the 5′ exon is not tethered, Brr2 activity may instead lead to dissociation of the U2 snRNP from the tri-snRNP (non-productive activation)12. b, The Prp3 model is superimposed on B1 (putative N-terminal region), B2 (helix α4) and B4 (C-terminal region) cryo-EM densities. The Prp3 ferredoxin-like fold (FER) and secondary structure elements are labelled. The black arrowhead indicates the region of Prp3 helix α4 that bends with different U2 snRNP positions (see a). For cryo-EM density nomenclature, see Extended Data Fig. 1f.
Extended Data Figure 6 Details of B complex proteins and U6 snRNA.
a, Fit of the Spp381 model to B6 (helices α1–α2) and B7 (helices α3–α4) cryo-EM densities. Helix α4 was modelled into weak density (B7) based on homology to the human and Chaetomium thermophilum crystal structures46. For cryo-EM density nomenclature, see Extended Data Fig. 1f. b, Fit of the Prp38 model to the B7 cryo-EM density. Helix α5 is shown below, revealing side-chain features in the density. c, Fit of the Snu23 model to B6 (helix α1) and B7 (remainder of Snu23) cryo-EM densities. Helix α2 is shown below, revealing side-chain features in the density. d, Composite cryo-EM density and fit of the U6 snRNA model. The B1, B2, B4, B5 and B7 densities are shown without (left) and with (right) the U6 snRNA model superimposed. U6 elements and the site of interaction with the U5 snRNA loop 1 are indicated. e, A weak density for pre-mRNA connects from the U6 snRNA ACAGAGA stem to the U2 SF3b-bound intron. The connecting density (map B2) is shown at intermediate (grey, threshold of 0.023) and low (light blue, threshold of 0.0173) thresholds. The U6 snRNA densities are shown as in d. The register of pre-mRNA near the U6 ACAGAGA stem is uncertain and was tentatively modelled based on complementarity with UBC4 pre-mRNA upstream and nearest to the 5′ exon, consistent with RNA crosslinking48. According to this register, approximately 40 pre-mRNA nucleotides separate U6- and SF3b-bound regions. The bottom right panel shows the fit of the pre-mRNA–U6 snRNA ACAGAGA stem–loop model to the unsharpened B7 density. For cryo-EM density nomenclature, see Extended Data Fig. 1f. f, Snu23 binds near the nucleotide-binding pocket of the Brr2 NHC, where it may influence Brr2 activity. Brr2 (pale cyan) and Snu23 (violet) models are superimposed on the B6 cryo-EM density, coloured as the underlying proteins. The RecA-1 and RecA-2 lobes of the Brr2 NHC are labelled, and an ADP nucleotide was modelled by aligning the NHC of human BRR2 bound to ADP73 (PDB code 4KIT) on the equivalent yeast Brr2 residues. The path of the Snu23 N terminus cryo-EM density is indicated, and is near to the Brr2 nucleotide-binding pocket. g, The U6 ACAGAGA stem is chaperoned by Dib1, Prp6, Prp8 and B complex proteins. Stabilization of the U6 ACAGAGA stem may facilitate tethering of pre-mRNA at its tip, whereas the U6 ACAGAGA box is buried in the stem. Surface models of Snu23 ZnF, Prp38 N terminus, Prp6 N terminus, Dib1, Prp8L and Prp8N domains, and Brr2 are shown and reveal a network of protein–RNA contacts to maintain the U6 ACAGAGA stem. h, Comparison of human tri-snRNP25 (PDB code 3JCR) and yeast B complex (this study) reveals that the Prp8N and Prp8EN domains serve as a platform for mutually exclusive binding of Prp28 and B complex proteins. Their Prp8N binding sites overlap and the altered location of the Prp8EN domain between the two complexes provides additional interfaces for either Prp28 or B complex proteins to bind. Movements compared to B complex in Brr2 and U4/U6 di-snRNP are indicated with arrows, and may occur after A complex association. The U2 snRNP is shown in grey to highlight tri-snRNP components, which are coloured as in Fig. 1. Comparison with Bact and C* structures20,21,22,23,24 further suggests that the Prp8N and Prp8EN domains serve as a general platform for step-specific splicing factors during the splicing cycle.
Extended Data Figure 7 U5 snRNA model, location of the U1 snRNP, and RNA secondary structure diagrams.
a, An improved model for U5 snRNA. A secondary structure diagram (left) and refined coordinate model (right) are shown, and the newly determined variable stem–loop II, stem III and stem IV are labelled together with known U5 snRNA elements. The long form of U5 snRNA is shown, where the short form ends with nucleotide 179. The grey boxes indicate regions not included in the model. Lines indicate Watson–Crick base pairs, and dots indicate G–U wobble base pairs. The U5 snRNA model was prepared by M. Wilkinson. b, Putative model of the RNA interaction network in the pre-B complex. The RNA network is unchanged compared to B complex, except for the interaction of the pre-mRNA 5′-exon with U1 snRNA74. Only loop 1 of U5 snRNA is shown. Lines indicate Watson–Crick base pairs, dots G–U wobble base pairs, stars denote non-canonical base pairs, and dotted lines putative nucleotide interactions. c, Putative location of the U1 snRNP. To gain insights into U1 snRNP location relative to U2 snRNP and tri-snRNP, we combined genetic, biochemical and structural observations. The U1 snRNP may bind between the human SF3a subunit SF3A1 (yeast Prp21)75, the Prp28-binding site25, Brr2 (ref. 76), and the U6 ACAGAGA stem. In B complex, the U1 snRNP may be destabilized owing to a steric clash with Brr2, which is likely to be repositioned compared to the pre-B complex, as in the human tri-snRNP structure25. In humans, the U1 snRNP may be further destabilized by a loss of A complex-specific proteins2. Brr2 repositioning may therefore serve as a checkpoint to signal the release of U1 snRNA from pre-mRNA. d, RNA secondary structure diagrams of regions modelled in B and Bact complex spliceosomes, using UBC4 pre-mRNA. The pre-mRNA substrate in Bact (ref. 21) is a mixture of cellular pre-mRNAs and its sequence is replaced by that of UBC4. The consensus nucleotides at the 5′SS and branch point sequence in yeast are shown in bold. Only loop 1 of U5 snRNA is shown. Lines indicate Watson–Crick base pairs, dots denote G–U wobble base pairs, stars denote non-canonical base pairs, and dotted lines putative nucleotide interactions. Highlighted are the branch-point adenosine, pre-mRNA 5′ and 3′ exons, and the U6 ACAGAGA box.
Extended Data Figure 8 Compositional and conformational changes during spliceosome activation.
a, List of RNA and protein components in B and Bact complex spliceosomes. During spliceosome activation, 22 proteins join the spliceosome, whereas 24 proteins (or 41 including the U1 snRNP) and U1 and U4 snRNAs are released. The U1 snRNP is indicated by a dashed line owing to its substoichiometric binding in B complex. RES complex proteins were not modelled in B complex, however, weak density is visible at the same binding site as in Bact (refs 20, 21), consistent with mass spectrometry data (ref. 9 and data not shown). b, Movement of the Prp8 RNase H domain (Prp8RH) between B and Bact complex spliceosomes. In B complex (left) regions of Prp3, Prp6 and Snu66 (residues 148–236) contact the Prp8RH and its β-hairpin (red). The Prp8 large domain (Prp8L) is shown as a surface and subunits are coloured as in Fig. 1. An arrow indicates the movement of Prp8RH to its Bact position21 (right; PDB code 5GM6), where it is stabilized by Hsh155, Prp45 (yellow) and Cwc22 (dark violet). B and Bact models were aligned on the Prp8L. c, Movements of the Prp8 switch loop and N-terminal domain between B and Bact complex spliceosomes. In B complex (left), an unassigned peptide (orange) binds the Prp8 switch loop, stabilizing it on the Prp8L domain. The Prp8 N-terminal domain (Prp8N) is shown as a surface (magenta) and binds the U6 snRNA 5′ stem. The Prp8 endonuclease (Prp8EN) domain is labelled. Arrows indicate the movements required to transition to Bact (PDB code 5GM6). The unassigned peptide is released in Bact (right) and Cwc21 (yellow) and Cwc22 (dark violet) bind in its stead to stabilize the new position of the Prp8 switch loop and the loaded pre-mRNA 5′ exon (light orange) in the exon channel. The repositioned Prp8N completes this channel. B and Bact models were aligned on the Prp8L domain. d, Movements of U2 3′ domain–SF3a subcomplexes between B and Bact complex spliceosomes. In B complex (left) the U2 3′ domain–SF3a subcomplexes are flexibly linked to SF3b and assume several positions relative to SF3b (compare Extended Data Fig. 4a). For comparison to Bact, the model for U2 3′ domain–SF3a (light green/grey) was rigid-body fitted into a low-pass filtered cryo-EM density of yeast Bact (EMDB code EMD-4099)20. This indicated that in Bact (right), the U2 3′ domain–SF3a complex is repositioned due to binding of NTC proteins Syf1 and Clf1 (yellow arch), to avoid a steric clash. The NTC subunits Syf1 and Clf1 may thus be positioned in Bact to bind the U2 3′ domain after the release of SF3a, during conversion to B* (Extended Data Fig. 8d). This repositioning is distinct from U2 3′ domain–SF3a conformations that are sampled in B complex. B and Bact models of the U2 snRNP were aligned on SF3b subunit Rse1. e, Movements of SF3b subunit Hsh155 and pre-mRNA between B and Bact complex spliceosomes. B and Bact (PDB code 5GM6) models were aligned on Hsh155, revealing small conformational changes in Hsh155 HEAT repeats, possibly owing to extended interaction of Hsh155 with Prp8, Snu17 and Prp45 (grey arches) in Bact. The U2–pre-mRNA branch helix is bound in the same manner in B and Bact complex spliceosomes. However, nucleotides downstream of the branch point (magenta) are bound differently, possibly due to movements in Hsh155. f, Structural differences between B and Bact complex spliceosomes suggest a model for activation. The ATP-dependent helicase Brr2 is positioned in B complex (left) and unwinds the U4/U6 duplex to release U4 snRNA and U4/U6 and B complex proteins (middle left). Their removal enables movements in Brr2, U2 and U5 snRNPs, and U6 snRNA to facilitate the loading of pre-mRNA, and formation of the U2/U6 catalytic centre (middle right). This intermediate is subsequently stabilized by NTC, NTR, splicing factors and Bact complex proteins to form Bact (ref. 21) (right; PDB code 5GM6). U5 snRNP (blue; U5 snRNA, light grey), U4/U6 di-snRNP (light yellow; U4 snRNA, yellow; U6 snRNA, red), U2 snRNP (U2 3′ domain, light green; SF3a, olive; SF3b, dark green), B complex proteins (shades of magenta), NTC, NTR and splicing factors (light yellow), and Bact complex proteins (shades of salmon) are indicated. The Bact position of U2 snRNP 3′ domain–SF3a was modelled as in Fig. 5. B and Bact spliceosomes were aligned on their Prp8L domain. Spliceosome activation intermediates are modelled.
Extended Data Figure 9 Data collection, refinement statistics and structure validation.
a, Cryo-EM data collection and refinement statistics of the B complex structure. Different regions of the composite B complex structure were refined into B1, B4, B5, B6 and B7 maps as described (see Methods). b, FSC between local cryo-EM map regions and corresponding regions of the refined coordinate models. Note that the FSC curve for the B-specific proteins correlates with the local resolution in this sub-region of the B7 density (4.0–5.0 Å; Extended Data Fig. 3e), below the nominal resolution (4.0 Å).
Supplementary information
Supplementary Figure
This file contains Supplementary Figure 1, comprising source data for protein and RNA gels. Supplementary Figure 1a, b, and c are the uncropped images for (a) Extended Data Fig. 1a, (b), Extended Data Fig. 1b, and (c) Extended Data Fig. c. (PDF 555 kb)
Supplementary Data
This file contains a pymol session of the B complex (PDB coordinate file: 5NRL). Colours as in Figure 1. (ZIP 5224 kb)
Structure of a pre-catalytic spliceosome
This video shows a 360 degree rotation of the B complex (PDB coordinate file: 5NRL). Colours as in Figure 1. (MOV 28942 kb)
Rights and permissions
About this article
Cite this article
Plaschka, C., Lin, PC. & Nagai, K. Structure of a pre-catalytic spliceosome. Nature 546, 617–621 (2017). https://doi.org/10.1038/nature22799
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature22799
This article is cited by
-
Cryo-EM analyses of dimerized spliceosomes provide new insights into the functions of B complex proteins
The EMBO Journal (2024)
-
The SMN complex drives structural changes in human snRNAs to enable snRNP assembly
Nature Communications (2023)
-
Estimating conformational landscapes from Cryo-EM particles by 3D Zernike polynomials
Nature Communications (2023)
-
OPUS-DSD: deep structural disentanglement for cryo-EM single-particle analysis
Nature Methods (2023)
-
Recruitment of a splicing factor to the nuclear lamina for its inactivation
Communications Biology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.