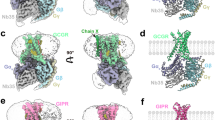
Abstract
The glucagon-like peptide-1 receptor (GLP-1R) and the glucagon receptor (GCGR) are members of the secretin-like class B family of G-protein-coupled receptors (GPCRs) and have opposing physiological roles in insulin release and glucose homeostasis1. The treatment of type 2 diabetes requires positive modulation of GLP-1R to inhibit glucagon secretion and stimulate insulin secretion in a glucose-dependent manner2. Here we report crystal structures of the human GLP-1R transmembrane domain in complex with two different negative allosteric modulators, PF-06372222 and NNC0640, at 2.7 and 3.0 Å resolution, respectively. The structures reveal a common binding pocket for negative allosteric modulators, present in both GLP-1R and GCGR3 and located outside helices V–VII near the intracellular half of the receptor. The receptor is in an inactive conformation with compounds that restrict movement of the intracellular tip of helix VI, a movement that is generally associated with activation mechanisms in class A GPCRs4,5,6. Molecular modelling and mutagenesis studies indicate that agonist positive allosteric modulators target the same general region, but in a distinct sub-pocket at the interface between helices V and VI, which may facilitate the formation of an intracellular binding site that enhances G-protein coupling.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
de Graaf, C. et al. Glucagon-like peptide-1 and its class B G protein-coupled receptors: a long march to therapeutic successes. Pharmacol. Rev. 68, 954–1013 (2016)
Cho, Y. M., Merchant, C. E. & Kieffer, T. J. Targeting the glucagon receptor family for diabetes and obesity therapy. Pharmacol. Ther. 135, 247–278 (2012)
Jazayeri, A. et al. Extra-helical binding site of a glucagon receptor antagonist. Nature 533, 274–277 (2016)
Rasmussen, S. G. et al. Crystal structure of the β2 adrenergic receptor–Gs protein complex. Nature 477, 549–555 (2011)
Carpenter, B., Nehmé, R., Warne, T., Leslie, A. G. & Tate, C. G. Structure of the adenosine A2A receptor bound to an engineered G protein. Nature 536, 104–107 (2016)
Scheerer, P. et al. Crystal structure of opsin in its G-protein-interacting conformation. Nature 455, 497–502 (2008)
Knudsen, L. B. et al. Small-molecule agonists for the glucagon-like peptide 1 receptor. Proc. Natl Acad. Sci. USA 104, 937–942 (2007)
Sloop, K. W. et al. Novel small molecule glucagon-like peptide-1 receptor agonist stimulates insulin secretion in rodents and from human islets. Diabetes 59, 3099–3107 (2010)
Chen, D. et al. A nonpeptidic agonist of glucagon-like peptide 1 receptors with efficacy in diabetic db/db mice. Proc. Natl Acad. Sci. USA 104, 943–948 (2007)
Yang, D. et al. Structural determinants of binding the seven-transmembrane domain of the glucagon-like peptide-1 receptor (GLP-1R). J. Biol. Chem. 291, 12991–13004 (2016)
Wootten, D. et al. The extracellular surface of the GLP-1 receptor is a molecular trigger for biased agonism. Cell 165, 1632–1643 (2016)
Underwood, C. R. et al. Crystal structure of glucagon-like peptide-1 in complex with the extracellular domain of the glucagon-like peptide-1 receptor. J. Biol. Chem. 285, 723–730 (2010)
Patterson, J. T., Li, P., Day, J. W., Gelfanov, V. M. & Dimarchi, R. D. A hydrophobic site on the GLP-1 receptor extracellular domain orients the peptide ligand for signal transduction. Mol. Metab. 2, 86–91 (2013)
Hollenstein, K. et al. Structure of class B GPCR corticotropin-releasing factor receptor 1. Nature 499, 438–443 (2013)
Siu, F. Y. et al. Structure of the human glucagon class B G-protein-coupled receptor. Nature 499, 444–449 (2013)
Wootten, D., Simms, J., Miller, L. J., Christopoulos, A. & Sexton, P. M. Polar transmembrane interactions drive formation of ligand-specific and signal pathway-biased family B G protein-coupled receptor conformations. Proc. Natl Acad. Sci. USA 110, 5211–5216 (2013)
Mann, R. J., Al-Sabah, S., de Maturana, R. L., Sinfield, J. K. & Donnelly, D. Functional coupling of Cys-226 and Cys-296 in the glucagon-like peptide-1 (GLP-1) receptor indicates a disulfide bond that is close to the activation pocket. Peptides 31, 2289–2293 (2010)
Zhang, H. et al. Structure of the full-length glucagon class B G protein-coupled receptor. Naturehttp://dx.doi.org/10.1038/nature22363 (this issue)
Zhang, D. et al. Two disparate ligand-binding sites in the human P2Y1 receptor. Nature 520, 317–321 (2015)
Srivastava, A. et al. High-resolution structure of the human GPR40 receptor bound to allosteric agonist TAK-875. Nature 513, 124–127 (2014)
Xiong, Y. et al. Discovery of a novel glucagon receptor antagonist N-[(4-(1S)-1-[3-(3, 5-dichlorophenyl)-5-(6-methoxynaphthalen-2-yl)-1H-pyrazol-1-yl]ethylphenyl)carbonyl]-β-alanine (MK-0893) for the treatment of type II diabetes. J. Med. Chem. 55, 6137–6148 (2012)
Lau, J. et al. New beta-alanine derivatives are orally available glucagon receptor antagonists. J. Med. Chem. 50, 113–128 (2007)
Guzman-Perez, A. et al. The design and synthesis of a potent glucagon receptor antagonist with favorable physicochemical and pharmacokinetic properties as a candidate for the treatment of type 2 diabetes mellitus. Bioorg. Med. Chem. Lett. 23, 3051–3058 (2013)
Hollenstein, K. et al. Insights into the structure of class B GPCRs. Trends Pharmacol. Sci. 35, 12–22 (2014)
Bueno, A. B. et al. Positive allosteric modulation of the glucagon-like peptide-1 receptor by diverse electrophiles. J. Biol. Chem. 291, 10700–10715 (2016)
Nolte, W. M. et al. A potentiator of orthosteric ligand activity at GLP-1R acts via covalent modification. Nat. Chem. Biol. 10, 629–631 (2014)
Spyridaki, K. et al. Structural–functional analysis of the third transmembrane domain of the corticotropin-releasing factor type 1 receptor: role in activation and allosteric antagonism. J. Biol. Chem. 289, 18966–18977 (2014)
Venkatakrishnan, A. J. et al. Molecular signatures of G-protein-coupled receptors. Nature 494, 185–194 (2013)
Mathi, S. K., Chan, Y., Li, X. & Wheeler, M. B. Scanning of the glucagon-like peptide-1 receptor localizes G protein-activating determinants primarily to the N terminus of the third intracellular loop. Mol. Endocrinol. 11, 424–432 (1997)
Cordomí, A. et al. Functional elements of the gastric inhibitory polypeptide receptor: Comparison between secretin- and rhodopsin-like G protein-coupled receptors. Biochem. Pharmacol. 96, 237–246 (2015)
Chun, E. et al. Fusion partner toolchest for the stabilization and crystallization of G protein-coupled receptors. Structure 20, 967–976 (2012)
Lv, X. et al. In vitro expression and analysis of the 826 human G protein-coupled receptors. Protein Cell 7, 325–337 (2016)
Cherezov, V. et al. Rastering strategy for screening and centring of microcrystal samples of human membrane proteins with a sub-10 microm size X-ray synchrotron beam. J. R. Soc. Interface 6 (suppl. 5), S587–S597 (2009)
Kabsch, W. Xds. Acta Crystallogr. D 66, 125–132 (2010)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007)
Adams, P. D . et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Vagin, A. A. et al. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D 60, 2184–2195 (2004)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
Smart, O. S. et al. Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER. Acta Crystallogr. D 68, 368–380 (2012)
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010)
Sastry, G. M., Adzhigirey, M., Day, T., Annabhimoju, R. & Sherman, W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 27, 221–234 (2013)
Zhu, K. et al. Docking covalent inhibitors: a parameter free approach to pose prediction and scoring. J. Chem. Inf. Model. 54, 1932–1940 (2014)
Klauda, J. B. et al. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B 114, 7830–7843 (2010)
Yu, W., He, X., Vanommeslaeghe, K. & MacKerell, A. D., Jr. Extension of the CHARMM General Force Field to sulfonyl-containing compounds and its utility in biomolecular simulations. J. Comput. Chem. 33, 2451–2468 (2012)
Pronk, S. et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 29, 845–854 (2013)
Hess, B. P-LINCS: a parallel linear constraint solver for molecular simulation. J. Chem. Theory Comput. 4, 116–122 (2008)
Bussi, G., Donadio, D. & Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 126, 014101 (2007)
Berendsen, H. J. C., Postma, J. P. M., van Gunsteren, W. F., DiNola, A. & Haak, J. R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81, 3684 (1984)
Mitternacht, S. FreeSASA: An open source C library for solvent accessible surface area calculations. F1000 Res. 5, 189 (2016)
Acknowledgements
This work was supported by the National Natural Science Foundation of China grants 31330019 (Z.-J.L.), 31500593 (G.S.), 81373463 and 81573479 (D.Y.), the National Health and Family Planning Commission grants 2012ZX09304-011, 2013ZX09401003-005, 2013ZX09507001 and 2013ZX09507-002 (M.-W.W.), Shanghai Science and Technology Development Fund 15DZ2291600 (M.-W.W.), and Ministry of Science and Technology of China grants 2014CB910400 (Z.-J.L.) and 2015CB910104 (Z.-J.L.), the Netherlands eScience Center (NLeSC)/NWO Enabling Technologies project: 3D-e-Chem, grant 027.014.201 (C.d.G.), the European Cooperation in Science and Technology Action CM1207 GLISTEN (C.d.G.), and National Key Research and Development Program of China 2016YCF0905902 (S.Z.). We thank the Cloning, Cell Expression and Protein Purification Core Facilities of iHuman Institute for their support. We thank L. Qu, Y. Feng and C. Ji for their technical assistance, Y. Liu, H. Tao, S. Qin, W. Shui, F. Ni, C. Zhang, J. Cheng, Q. Zhao and V. Cherezov for discussions or contributions at the early stages of this project, and A. Walker and S. Reedtz-Runge for critical review of the manuscript. We thank the Shanghai Municipal Government, ShanghaiTech University and GPCR Consortium for financial support. The synchrotron radiation experiments were performed at the BL41XU of Spring-8 with approval of the Japan Synchrotron Radiation Research Institute (JASRI) (proposal no. 2016B2708 and 2016B2724), and beamline BL17U1 (Shanghai Synchrotron Radiation Facility [SSRF], China). Computational resources were supported by the Shanghai Supercomputer Center, and the Supercomputing Center of ShanghaiTech University.
Author information
Authors and Affiliations
Contributions
G.S., C.d.G. and M.A.H. designed constructs for crystallization. G.S., Y.W., S.J. and K.L. expressed, characterized and screened constructs and ligands for crystallization. G.S., Y.W., S.J. and F.W. purified and crystallized the receptor, optimized crystallization conditions and grew the crystals. G.S., Z.-J.L., M.A.H. and G.W.H. collected diffraction data and solved and refined the structure. G.S., D.Y. and C.d.G. designed and analysed the receptor mutagenesis studies. D.Y., X.C., A.D. and G.L. expressed the receptor, and performed the mutagenesis, functional and ligand-binding assays. Q.Z., Y.W., C.d.G. and S.Z. constructed the GLP-1R–agonist PAM model and performed and analysed MD-simulations on wild-type and mutant GLP-1R. B.W. provided advice on construct design. R.C.S. conceived of the project. R.C.S., M.-W.W., and Z.-J.L. were responsible for the overall project management and edited the manuscript. G.S., C.d.G. and M.A.H. wrote the manuscript with discussions and improvements from D.L., L.Y. and J.L.
Corresponding authors
Ethics declarations
Competing interests
J.L. is an employee of Novo Nordisk, a pharmaceutical company focused on GLP-1R for type 2 diabetes. R.C.S. is a founder and board member of Bird Rock Bio, a company focused on GPCR therapeutic antibodies. The remaining authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks G. Lebon, T. W. Schwartz and C. Siebold for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 GLP-1R crystallization and structure determination.
a, Schematic diagram of GLP-1R TMD construct (residues 128–431). Thermostabilizing mutations (green) are S193C, I196F, S225A, M233C, S271A, I317C, G318I, K346A, C347F and G361C. The single most conserved residue among class B GPCRs in each transmembrane helix (designated X.50b in the Wootten residue numbering scheme16, in which ‘X’ is the transmembrane helix number) is indicated in bold. Disordered residues in the structure are shown with a brown background. Engineered ECL1 linker residues are shown as dashed circles. The endogenous and engineered disulfide bonds are shown with solid and dashed orange lines, respectively. b, Thermostability assay (CPM) of representative mutations. Constructs that include the disulfide bond (I317C/G361C) are much sharper in curve transition compared to the control (no mutation, black line). c, Thermostability assay of the apo state protein (crystallization construct) and with PF-06372222 or NNC0640. In b and c, the dotted and solid lines represent the original and fitted curves, respectively. d, Representative crystals of GLP-1R–PF-06372222 in lipidic cubic phase. e, Crystal packing of GLP-1R–PF-06372222, with the two molecules per asymmetric unit coloured green and cyan, respectively. f, The |Fo| − |Fc| omit maps of PF-06372222 and NNC0640 contoured at 3.0σ.
Extended Data Figure 2 GLP-1R interhelical interaction network.
a, Overview of representative interactions. b, The conserved disulfide bond between C2263.29b and C296ECL2. c, Hydrogen-bond interactions between helices I and VII. d–f, Interaction network between helices III and VI (d), helices III, IV and V (e), and helices II, III and IV (f). This figure can be compared with figure 4 in ref. 14 (CRF1R) and figure 3 in ref. 15 (GCGR).
Extended Data Figure 3 Statistics of buried and exposed surface areas of 86 crystallized GPCR ligands.
a, Scatter plot of current crystallized ligands. x axis is the surface area buried by receptor; y axis is the ratio of exposed area to buried area. All surface areas were calculated based on the crystal structures using freeSASA48. PDBs with multiple chains are averaged in the plot. b–d, Current binding modes of GPCR allosteric modulators. For clarity, modulators that bind to similar region were arranged in the same cartoon.
Extended Data Figure 4 Effects of binding pocket mutations on potency of NAMs.
a–l, Dose-dependent inhibition curves of NAMs on wild-type GLP-1R (a) or GLP-1R mutants (b–j), as well as wild-type GCGR (k) or the F3456.36bC mutant (l). m, Summary of half-maximum inhibitory concentration (pIC50) values of NAMs on the above constructs. Relative expression levels of mutated constructs were evaluated by comparing to that of wild-type GLP-1R. Experiments were repeated at least three times and error bars represent s.e.m. of quadruplicate measurements. NA, not available.
Extended Data Figure 5 C3476.36bF stabilizes the interaction interface between GLP-1R and NAMs.
Superposition of PF-06372222-bound crystal structure (containing the C3476.36bF mutation), with crystal structure based molecular dynamic simulations of F347 shown in blue (500 ns) and of simulations C347 (6.36b mutated back to cysteine) shown in pink (500 ns). PF-06372222 and key residues are shown as sticks. For clarity, only the backbone of the crystal structure is shown (grey). In the simulation of F347, the ligand adopts the same orientation as in the crystal structure, whereas the orientation of the ligand in the simulation of C347 is varied during the simulation process (the trifluoromethyl-pyrazole group in particular).
Extended Data Figure 6 Binding of GLP-1 and exendin-4(9-39) to representative constructs.
a, b, The four representative constructs used in Fig. 3d are tested for their binding properties with full agonist GLP-1 (a), and fragment antagonist exendin-4(9-39) that targets the extracellular domain (b). Experiments were repeated at least three times and error bars represent s.e.m. of duplicate measurements. NB, no binding.
Extended Data Figure 7 Conformational changes revealed by molecular dynamics simulation.
a, Comparison of compound 2 (grey) docked to the GLP-1R crystal structure, after 500 ns molecular dynamics simulation (compound 2 in cyan), and molecular dynamics simulation of PF-06372222-bound GLP-1R (PF-06372222 in pink). b, The hydrogen-bond interaction network between residues associated with the ionic lock observed in the GLP-1R crystal structure14,15,24. c, In molecular dynamics simulation of the PF-06372222-bound GLP-1R crystal structure, the ionic lock hydrogen bond network is preserved. d, Molecular dynamics simulation of compound 2 covalently bound to wild-type GLP-1R reveals unwinding of the N-terminal end of helix VI (IKC6.36bRL, coloured orange) and re-organization of the ionic lock interaction network. Unwinding of helix VI disrupts the ionic lock interactions between R3486.37b and E4087.63b and destabilizes the hydrogen-bond interaction between H1802.50b and E2473.50b. These conformational changes allow R1762.46b to hydrogen bond with E2473.50b, and reinforce the hydrogen bond between T3536.44b and Y4027.57b compared to the PF-06372222-bound GLP-1R molecular dynamics simulation. e–g, Intracellular views (surface representation) of b–d. h, Hydrogen-bond interactions between key residues during simulations in c and d. Hydrogen bonds were determined with the g_hbond program in the Gromacs45, using a hydrogen bond distance cut-off of 3.5 Å and angle cut-off of 120°–240°. i, Six residues around the intracellular ionic lock of GLP-1R (H1802.50b, L2513.54b, L3496.40b, S3506.41b, T3536.44b and Y4027.57b) were selected to calculate the solvent-accessible surface areas (SASA) using the program freeSASA49. Compared to the crystal structure or simulation of PF-06372222, these residues (marked red in e–g) were exposed to solvent by 40–100 Å2 in the simulation of compound 2 (g, i). In e and f, these residues are buried, and thus are not visible, while in g, exposure of these residues provides space for the G protein to bind.
Extended Data Figure 8 Effects of the PAM binding pocket mutations on potency of compound 2.
a–c, Wild-type GLP-1R and 18 mutants (including 2 double mutants) were compared for their effects on potency of compound 2. Curves were coloured based on the same criteria as in Fig. 4 and codes were ranked based on their pEC50 (negative logarithm of the half-maximum effective concentration (EC50)) values (listed in the tables). Experiments were repeated at least three times and error bars represent s.e.m. of quadruplicate measurements.
Extended Data Figure 9 The R176Q mutation decreases the potency of GLP-1R, but does not affect its binding capacity with GLP-1.
a, b, Comparison of wild-type GLP-1R and the R1762.46bQ mutant by GLP-1 binding assay (a) and functional cAMP accumulation assay (b). cAMP accumulation assay is conducted as described in the text, and the binding assay was carried out using radiolabelled GLP-1 as a tracer and competing it with a serial dilution of unlabelled GLP-1. Experiments were repeated three times and error bars represent s.e.m. of duplicate (binding assay) or quadruplicate (cAMP assay) measurements.
Rights and permissions
About this article
Cite this article
Song, G., Yang, D., Wang, Y. et al. Human GLP-1 receptor transmembrane domain structure in complex with allosteric modulators. Nature 546, 312–315 (2017). https://doi.org/10.1038/nature22378
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature22378
This article is cited by
-
G protein-coupled receptors (GPCRs): advances in structures, mechanisms, and drug discovery
Signal Transduction and Targeted Therapy (2024)
-
A framework for Frizzled-G protein coupling and implications to the PCP signaling pathways
Cell Discovery (2024)
-
GPR161 structure uncovers the redundant role of sterol-regulated ciliary cAMP signaling in the Hedgehog pathway
Nature Structural & Molecular Biology (2024)
-
Structure-based drug discovery of a corticotropin-releasing hormone receptor 1 antagonist using an X-ray free-electron laser
Experimental & Molecular Medicine (2023)
-
GLP-1R signaling neighborhoods associate with the susceptibility to adverse drug reactions of incretin mimetics
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.