Abstract
The mechanistic target of rapamycin (mTOR) has a key role in the integration of various physiological stimuli to regulate several cell growth and metabolic pathways1. mTOR primarily functions as a catalytic subunit in two structurally related but functionally distinct multi-component kinase complexes, mTOR complex 1 (mTORC1) and mTORC2 (refs 1, 2). Dysregulation of mTOR signalling is associated with a variety of human diseases, including metabolic disorders and cancer1. Thus, both mTORC1 and mTORC2 kinase activity is tightly controlled in cells. mTORC1 is activated by both nutrients3,4,5,6 and growth factors7, whereas mTORC2 responds primarily to extracellular cues such as growth-factor-triggered activation of PI3K signalling8,9,10. Although both mTOR and GβL (also known as MLST8) assemble into mTORC1 and mTORC2 (refs 11, 12, 13, 14, 15), it remains largely unclear what drives the dynamic assembly of these two functionally distinct complexes. Here we show, in humans and mice, that the K63-linked polyubiquitination status of GβL dictates the homeostasis of mTORC2 formation and activation. Mechanistically, the TRAF2 E3 ubiquitin ligase promotes K63-linked polyubiquitination of GβL, which disrupts its interaction with the unique mTORC2 component SIN1 (refs 12, 13, 14) to favour mTORC1 formation. By contrast, the OTUD7B deubiquitinase removes polyubiquitin chains from GβL to promote GβL interaction with SIN1, facilitating mTORC2 formation in response to various growth signals. Moreover, loss of critical ubiquitination residues in GβL, by either K305R/K313R mutations or a melanoma-associated GβL(ΔW297) truncation, leads to elevated mTORC2 formation, which facilitates tumorigenesis, in part by activating AKT oncogenic signalling. In support of a physiologically pivotal role for OTUD7B in the activation of mTORC2/AKT signalling, genetic deletion of Otud7b in mice suppresses Akt activation and Kras-driven lung tumorigenesis in vivo. Collectively, our study reveals a GβL-ubiquitination-dependent switch that fine-tunes the dynamic organization and activation of the mTORC2 kinase under both physiological and pathological conditions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Zoncu, R., Efeyan, A. & Sabatini, D. M. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 12, 21–35 (2011)
Aylett, C. H. et al. Architecture of human mTOR complex 1. Science 351, 48–52 (2016)
Sancak, Y. et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320, 1496–1501 (2008)
Zoncu, R. et al. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H+-ATPase. Science 334, 678–683 (2011)
Bar-Peled, L., Schweitzer, L. D., Zoncu, R. & Sabatini, D. M. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell 150, 1196–1208 (2012)
Bar-Peled, L. et al. A tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science 340, 1100–1106 (2013)
Inoki, K., Li, Y., Xu, T. & Guan, K. L. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 17, 1829–1834 (2003)
Guertin, D. A. & Sabatini, D. M. Defining the role of mTOR in cancer. Cancer Cell 12, 9–22 (2007)
Liu, P. et al. Sin1 phosphorylation impairs mTORC2 complex integrity and inhibits downstream Akt signalling to suppress tumorigenesis. Nat. Cell Biol. 15, 1340–1350 (2013)
Liu, P. et al. PtdIns(3,4,5)P3-dependent activation of the mTORC2 kinase complex. Cancer Discov. 5, 1194–1209 (2015)
Kim, D. H. et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110, 163–175 (2002)
Yang, Q., Inoki, K., Ikenoue, T. & Guan, K. L. Identification of Sin1 as an essential TORC2 component required for complex formation and kinase activity. Genes Dev. 20, 2820–2832 (2006)
Jacinto, E. et al. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell 127, 125–137 (2006)
Frias, M. A. et al. mSin1 is necessary for Akt/PKB phosphorylation, and its isoforms define three distinct mTORC2s. Curr. Biol. 16, 1865–1870 (2006)
Guertin, D. A. et al. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCα, but not S6K1. Dev. Cell 11, 859–871 (2006)
Komander, D. & Rape, M. The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 (2012)
Ikeda, F., Crosetto, N. & Dikic, I. What determines the specificity and outcomes of ubiquitin signaling? Cell 143, 677–681 (2010)
Linares, J. F. et al. K63 polyubiquitination and activation of mTOR by the p62-TRAF6 complex in nutrient-activated cells. Mol. Cell 51, 283–296 (2013)
Jin, G. et al. Skp2-mediated RagA ubiquitination elicits a negative feedback to prevent amino-acid-dependent mTORC1 hyperactivation by recruiting GATOR1. Mol. Cell 58, 989–1000 (2015)
Deng, L. et al. The ubiquitination of rag A GTPase by RNF152 negatively regulates mTORC1 activation. Mol. Cell 58, 804–818 (2015)
Inoki, K., Li, Y., Zhu, T., Wu, J. & Guan, K. L. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat. Cell Biol. 4, 648–657 (2002)
Guertin, D. A. et al. mTOR complex 2 is required for the development of prostate cancer induced by Pten loss in mice. Cancer Cell 15, 148–159 (2009)
Mevissen, T. E. et al. OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell 154, 169–184 (2013)
Hussain, S. et al. Ubiquitin hydrolase UCH-L1 destabilizes mTOR complex 1 by antagonizing DDB1-CUL4-mediated ubiquitination of raptor. Mol. Cell. Biol. 33, 1188–1197 (2013)
Mevissen, T. E. et al. Molecular basis of Lys11-polyubiquitin specificity in the deubiquitinase Cezanne. Nature 538, 402–405 (2016)
Bremm, A., Freund, S. M. & Komander, D. Lys11-linked ubiquitin chains adopt compact conformations and are preferentially hydrolyzed by the deubiquitinase Cezanne. Nat. Struct. Mol. Biol. 17, 939–947 (2010)
Gupta, S. et al. Binding of ras to phosphoinositide 3-kinase p110alpha is required for ras-driven tumorigenesis in mice. Cell 129, 957–968 (2007)
Castellano, E. et al. Requirement for interaction of PI3-kinase p110α with RAS in lung tumor maintenance. Cancer Cell 24, 617–630 (2013)
Hu, H. et al. OTUD7B controls non-canonical NF-κB activation through deubiquitination of TRAF3. Nature 494, 371–374 (2013)
Pareja, F. et al. Deubiquitination of EGFR by Cezanne-1 contributes to cancer progression. Oncogene 31, 4599–4608 (2012)
Yuan, L. et al. Deubiquitylase OTUD3 regulates PTEN stability and suppresses tumorigenesis. Nat. Cell Biol. 17, 1169–1181 (2015)
Bremm, A., Moniz, S., Mader, J., Rocha, S. & Komander, D. Cezanne (OTUD7B) regulates HIF-1α homeostasis in a proteasome-independent manner. EMBO Rep. 15, 1268–1277 (2014)
Wei, W., Jobling, W. A., Chen, W., Hahn, W. C. & Sedivy, J. M. Abolition of cyclin-dependent kinase inhibitor p16Ink4a and p21Cip1/Waf1 functions permits Ras-induced anchorage-independent growth in telomerase-immortalized human fibroblasts. Mol. Cell. Biol. 23, 2859–2870 (2003)
Gan, W. et al. SPOP promotes ubiquitination and degradation of the ERG oncoprotein to suppress prostate cancer progression. Mol. Cell 59, 917–930 (2015)
Ordureau, A. et al. Defining roles of PARKIN and ubiquitin phosphorylation by PINK1 in mitochondrial quality control using a ubiquitin replacement strategy. Proc. Natl Acad. Sci. USA 112, 6637–6642 (2015)
Ordureau, A. et al. Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis. Mol. Cell 56, 360–375 (2014)
Phu, L. et al. Improved quantitative mass spectrometry methods for characterizing complex ubiquitin signals. Mol. Cell. Proteomics. 10, M110.003756 (2011)
MacLean, B . et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 (2010)
Huttlin, E. L. et al. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189 (2010)
Beausoleil, S. A., Villén, J., Gerber, S. A., Rush, J. & Gygi, S. P. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 24, 1285–1292 (2006)
Kim, W. et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–340 (2011)
Ran, F. A. et al. Genome engineering using the CRISPR–Cas9 system. Nat. Protocols 8, 2281–2308 (2013)
Sarbassov, D. D. et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 14, 1296–1302 (2004)
Johnson, L. et al. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature 410, 1111–1116 (2001)
Gyo˝rffy, B., Surowiak, P., Budczies, J. & Lánczky, A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One 8, e82241 (2013)
Acknowledgements
We thank T. Jacks and the NCI Mouse Repository for providing the KrasLA2 mice. We thank P. P. Pandolfi (Harvard), B. D. Manning (Harvard), and A. Toker (Harvard) for their insightful suggestions and critiques during the preparation of this manuscript. We also thank all Wei laboratory members for critical reading of the manuscript. W.W. is a LLS research scholar. P.L. is supported by ROOCA181342. W.G. is supported by 1K99CA207867. A.O. was supported by an Edward R. and Anne G. Lefler Center postdoctoral fellowship. This work was supported by NIH grants (W.W., R01CA177910 and R01GM094777; S.-C.S., R37AI064639 and R01GM084459; J.W.H., AG011085 and GM095567) and the National Natural Science Foundation of China (B.W., 81472294, L.Z., 81521064).
Author information
Authors and Affiliations
Contributions
B.W., Z.J., P.L., S.-C.S. and W.W. designed the research. B.W., Z.J., A.O., D.J., P.L., W.G., J.G., J.Z., B.J.N., X.D. and X.C. performed experiments and/or analysed data. S.-C.S. and W.W. supervised the study. X.B., L.Z. and J.W.H. provided critical reagents. B.W., Z.J., P.L., S.-C.S. and W.W. interpreted data and wrote the manuscript. All authors commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks D. Fruman and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
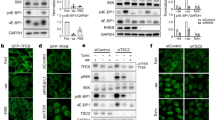
Extended Data Figure 1 The polyubiquitination status of GβL and mTORC2 kinase formation is regulated by upstream physiological stimuli.
a-c, The polyubiquitination status of GβL is reduced in response to growth factor stimulation. Immunoblot (IB) analysis of whole-cell lysates (WCL) or Ni-NTA pull-down products under denaturing conditions derived from HEK293 cells transfected with His–Ub together with (a, c) or without (b) HA–GβL vector. Cells were serum starved for 16 h and treated with insulin (100 nM) or Dulbecco’s phosphate-buffered saline (DPBS) for 30 min (a, c), or EGF (100 ng ml−1) for 16 min (b) before harvesting. d, e, Unlike GβL, polyubiquitination of RPTOR is minimally affected by growth signalling induction. IB analysis of WCL or Ni-NTA pull-down products under denaturing conditions derived from HEK293 cells transfected with HA–RPTOR and His–Ub constructs. Thirty-two hours post-transfection, cells were subjected to serum starvation for 16 h, and then exposed to insulin (100 nM) (d) or EGF (100 ng ml−1) (e) at indicated time points before harvesting. f, Quantification results from three independent experiments showing fold differences of endogenous GβL binding to SIN1, RICTOR and Rptor in cells in response to insulin stimulation, as indicted in Fig. 1c. Data are mean ± s.d. The intensity of each blot generated using ImageJ software were analysed by two-tailed paired Student’s t-test, *P < 0.05, **P < 0.01. g, The assembly of endogenous mTORC1 and mTORC2 responds to physiological growth factor stimulation. IB analysis of GβL immunoprecipitate or WCL derived from HEK293 cells serum starved for 16 h, then stimulated with EGF (100 ng ml−1), and lysed at indicated time points using CHAPS buffer for immunoprecipitation and IB analysis. Rabbit IgG antibody was used as negative control for GβL immunoprecipitation. For quantification analysis, levels were arbitrarily set at 1.0 at the 0 min time point. h–j, Growth factor stimulation enhances mTORC2, but inhibits mTORC1 formation at early time points. IB analysis of HA immunoprecipitate or WCL derived from HEK293 cells transfected with HA–mTOR (h), HA–RPTOR (i), or HA–RICTOR (j) constructs. Thirty-two hours post-transfection, cells were serum starved for 16 h, then stimulated with insulin (100 nM), and lysed at indicated time points using CHAPS buffer for immunoprecipitation and IB analysis. k, Polyubiquitination of GβL inversely correlates with mTORC2 integrity and activity in response to EGF stimulation. IB analysis of WCL or Ni-NTA pull-down products under denaturing conditions derived from HEK293 cells transfected with HA–GβL and His–Ub constructs. Thirty-two hours post-transfection, cells were subjected to serum starvation for 16 h, then treated with EGF (100 ng ml−1) for indicated time points, lysed for HA immunoprecipitation or His pull-down assays and IB analysis. l, Transient insulin stimulation increases the relative abundance of endogenous mTORC2, but reduces mTORC1 formation in cells. HEK293 cells were subjected to serum starvation for 16 h and treated with or without insulin (100 nM) for 15 min, and lysed using CHAPS buffer. WCL was filtered and run through an FPLC Superdex 200 column. Five hundred microlitres of elute was collected for each fraction and a 1/20 volume of each fraction was resolved on SDS–PAGE and subjected to IB analysis. m, GβL polyubiquitination linkage was examined by transfecting His-tagged wild-type (WT) and indicated KR ubiquitin mutants together with HA–GβL into HEK293 cells, followed by IB analysis of Ni-NTA pull-down products and WCL. n, Polyubiquitination of GβL could largely be detected in cells transfected with wild-type ubiquitin and lysine-63-only ubiquitin (K63 only), but not lysine-48-only ubiquitin mutant (K48 only) constructs. IB analysis of Ni-NTA pull-down products and WCL derived from HEK293 cells transfected with HA–GβL and wild-type His–Ub or mutant constructs as indicated. For uncropped gels, see Supplementary Fig. 1.
Extended Data Figure 2 The TRAF2 E3 ligase catalyses K63-linked polyubiquitination of GβL and inhibits the activity of mTORC2 kinase in cells.
a–c, GβL specifically interacts with TRAF2 in cells. IB analysis of immunoprecipitate and WCL derived from HEK293 cells transfected with indicated constructs. d, e, Polyubiquitination of GβL in HEK293 cells was examined by transfecting GβL, various indicated E3 ligases and His–Ub constructs, followed by IB analysis of WCL and Ni-NTA pull-down products under denaturing conditions. f, TRAF2, but not TRAF6, promotes polyubiquitination of endogenous GβL in cells. IB analysis of WCL or Ni-NTA pull-down products under denaturing conditions derived from HEK293 cells transfected indicated constructs. g, h, Genetic deletion of Traf2 attenuates polyubiquitination of GβL in cells. IB analysis of WCL or Ni-NTA pull-down products under denaturing conditions derived from Traf2+/+ and Traf2−/− MEFs that were transfected with (g) or without (f) HA–GβL, along with His–Ub vector. i, TRAF2 promotes polyubiquitination of GβL in an E3 ligase activity-dependent manner. IB analysis of WCL or Ni-NTA pull-down products under denaturing conditions derived from Traf2−/− MEFs transfected with HA–GβL and His–Ub, along with wild-type or the activity-deficient TRAF2 (ΔRING) mutant construct. j, TRAF2 promotes K63-linked polyubiquitination of GβL in cells. IB analysis of WCL or Ni-NTA pull-down products under denaturing conditions derived from HEK293 cells transfected with HA–GβL and His–Ub construct, or His-tagged ubiquitin mutants containing only lysine 63 (K63 only) or no lysine residue (K0). k, Genetic deletion of Traf2 does not affect the half-life of endogenous GβL protein in cells. IB analysis of WCL derived from Traf2+/+ and Traf2−/− MEFs treated with 100 μg ml−1 cycloheximide (CHX) for indicated time points before harvesting. l, A schematic diagram showing the evolutionarily conserved consensus TRAF2-binding motif within GβL. m, Mutating the TRAF2 consensus motif abolishes GβL binding to TRAF2 in cells. IB analysis of WCL and immunoprecipitate from HEK293 cells transfected with TRAF2, together with GβL or GβL(PEAA) constructs. n, Mutating the TRAF2 consensus motif largely abolishes GβL polyubiquitination in cells. Polyubiquitination status of GβL in HEK293 cells was examined by transfecting HA-tagged GβL or GβL(PEAA) with His–Ub constructs, followed by IB analysis of WCL and Ni-NTA pull-down products under denaturing conditions. o, Genetic deletion of Traf2 attenuates polyubiquitination of GβL in cells. IB analysis of WCL or Ni-NTA pull-down products under denaturing conditions derived from Traf2+/+ and Traf2−/− MEFs transfected with HA–GβL and His–Ub. Thirty-two hours post-transfection, cells were subjected to serum starvation for 16 h and exposed to EGF (100 ng ml−1) for indicated time points before harvesting. p, Loss of Traf2 leads to elevated Akt phosphorylation in response to EGF. IB analysis of WCL derived from Traf2+/+ and Traf2−/− MEFs that were subjected to serum starvation for 16 h and treated with EGF (100 ng ml−1) for indicated time points before harvesting. For quantification analysis, levels are normalized to total Akt or S6k1, respectively, and arbitrarily set at 1.0 at the 8 min time point of Traf2+/+ MEFs. q, TRAF2 inhibits Akt(pSer473) in an E3 ligase activity-dependent manner. IB analysis of WCL derived from Traf2−/− MEFs transfected with Flag-tagged wild-type TRAF2 or the activity-deficient TRAF2(ΔRING) constructs. r, Genetic deletion of Traf2 minimally affects Akt ubiquitination in cells. IB analysis of WCL or Ni-NTA pull-down products under denaturing conditions derived from Traf2+/+ and Traf2−/− MEFs transfected with HA–Akt1 and His–Ub. s, Inhibition of mTOR kinase activity does not significantly affect polyubiquitination of GβL in cells. IB analysis of WCL or Ni-NTA pull-down products under denaturing conditions derived from Traf2−/− MEFs transfected with HA–GβL and His–Ub constructs. Thirty-two hours post-transfection, cells were treated with or without Torin-1 (50 nM) for 6 h before harvesting. t, Activation of TNF receptor signalling does not significantly regulate polyubiquitination of GβL in cells. IB analysis of WCL or Ni-NTA pull-down products under denaturing conditions derived from mouse B cell lymphoma cell line A20 cells that were transfected with HA–GβL and His–Ub constructs. Thirty-two hours post-transfection, cells were treated with or without TNFα (50 ng ml−1) for indicated time points before harvesting. For uncropped gels, see Supplementary Fig. 1.
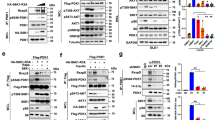
Extended Data Figure 3 TRAF2 promotes polyubiquitination of GβL at the WD7 motif, a specific SIN1-interacting domain, to impair the assembly of mTORC2 kinase.
a, b, GβL utilizes different WD40 motifs to interact with the two distinct mTOR complexes. Specifically, the WD6 motif is the major GβL domain interacting with the unique mTORC1 subunit, RPTOR, while the WD7 motif primarily binds a mTORC2-specific subunit, SIN1 in cells. IB analysis of WCL and GST pull-down products derived from HEK293 cells transfected with HA–RPTOR (a) or HA–SIN1 (b) constructs, together with indicated GST–GβL mammalian expression vectors. c, A schematic illustration showing specific binding of RPTOR to the WD6 domain of GβL to be incorporated into mTORC1, and binding of SIN1 to the WD7 motif of GβL to promote mTORC2 formation. d, A schematic model showing the experimental procedures of a sequential pull-down assay, which is performed to detect the existence of GβL ubiquitin moieties in either GST-purified mTORC1 or mTORC2. e, Detection of ubiquitinated GβL species in GST-purified mTORC1, but not mTORC2. HEK293 cells were transfected with either GST–SIN1 or GST–Rptor expression constructs, together with His–GβL and HA–Ub vectors. Forty-eight hours post-transfection, cells were harvested in CHAPS buffer to specifically pull down the intact mTORC1 or mTORC2 from cell lysates with GST-conjugated beads. The GST pull-down products were eluted using glutathione (GSH)-containing buffer and the elutes were subjected to second round of His pull-down assays in denaturing condition. Afterwards, the resulting samples were resolved on SDS–PAGE and subjected to IB analysis to detect the ubiquitination status of mTORC1- and mTORC2-associated GβL, respectively. f, GβL is largely ubiquitinated in GST-purified mTORC1, but not mTORC2. HEK293 cells were transfected with either GST–SIN1 or GST–Rptor constructs, together with His–Ub vector. Following a similar protocol as described in e, His pull-down products and WCL were subjected to IB analysis of GβL ubiquitin moieties in either mTORC1 or mTORC2. g, Quantification results from three independent experiments showing fold differences of endogenous GβL binding to Sin1, Rictor and Rptor in Traf2+/+ and Traf2−/− MEFs, as indicated in Fig. 2d (mean ± s.d., *P < 0.05, **P < 0.01, paired Student’s t-test). h, IB analysis of WCL and HA immunoprecipitate derived from Traf2+/+ and Traf2−/− MEFs transfected with HA–GβL or empty vector (EV) as a negative control. i, Deletion of Traf2 increases the formation of endogenous mTORC2, meanwhile reduces endogenous mTORC1 formation. Traf2+/+ and Traf2−/− MEFs in normal culture medium were lysed using CHAPS buffer. WCL was fractionated through an FPLC Superdex 200 column. Five hundred microlitres of eluate was collected for each fraction and a 1/20 volume of each fraction was resolved on SDS–PAGE for IB analysis. j, TRAF2 promotes GβL interaction with RPTOR, but inhibits GβL–SIN1 binding in an E3-ligase-activity-dependent manner. IB analysis of WCL and HA immunoprecipitate derived from HEK293 cells transfected with HA–GβL, Myc–RPTOR, Flag–SIN1, together with increasing doses of wild-type TRAF2 or the activity-deficient TRAF2(ΔRING) mutant construct. For uncropped gels, see Supplementary Fig. 1.
Extended Data Figure 4 Deficiency in GβL ubiquitination at K305/K313 of the WD7 domain promotes mTORC2 formation and kinase activity in cells.
a, A schematic diagram showing two evolutionarily conserved lysine residues (K305 and K313) within the WD7 domain of GβL. b, Mass spectrometry analysis to identify K305 as one of the major GβL ubiquitination residues within the WD7 domain of GβL in cells. HEK293 cells were transfected with CMV-GST-GβL and ubiquitin expression constructs. Forty-eight hours post-transfection, cells were lysed using Triton buffer for GST pull down. The GST pull-down products were eluted with GSH-containing buffer and then subjected to mass spectrometry analysis of ubiquitination sites. The recovered GβL peptide and the ubiquitination site (K305) were highlighted in red. c, Mutating key lysine residues within the WD7 domain of GβL abolishes GβL ubiquitination in cells. Polyubiquitination of GβL in HEK293 cells was examined by transfecting the indicated HA–GβL constructs with His–Ub, followed by IB analysis of WCL and Ni-NTA pull-down products under denaturing conditions. d, Sequencing of PCR products from genomic DNA demonstrates the introduction of A to G substitution at the codon encoding K305 and K313 of GβL gene in a GβLKRKR knock-in HEK293 cell line generated by CRISPR-mediated gene editing. e, Loss of GβL ubiquitination leads to elevated GβL binding to mTORC2 components, and reduced GβL binding to mTORC1 components in cells. IB analysis of WCL and HA immunoprecipitate derived from HEK293 cells transfected with indicated GβL plasmids. f, GβLKRKR knock-in cells, compared to wild-type cells, display an increased formation of endogenous mTORC2 and a reduced formation of endogenous mTORC1. HEK293 cells harbouring wild-type GβL, or ubiquitination-deficient GβLKRKR were lysed using CHAPS buffer. The WCL was run through an FPLC Superdex 200 column to collect fractionated cell eluates. A 1/20 volume of each fraction was subjected to IB analysis. g, Loss of ubiquitination of endogenous GβL promotes mTORC2 formation in CHAPS and EBC buffer, but not Triton buffer. IB analysis of GβL immunoprecipitate derived from GβLKRKR knock-in or wild-type HEK293 cells lysed using indicated buffers for immunoprecipitation. The resulting samples were resolved on SDS–PAGE for IB analysis. IB result of WCL prepared using Triton buffer were shown as input. h, Mutating the specific TRAF2 binding site in GβL promotes GβL integration into mTORC2, but inhibits its integration into mTORC1. IB analysis of WCL and HA immunoprecipitate derived from HEK293 cells transfected with HA–GβL or the TRAF2-non-interacting GβL(PEAA) mutant construct. i, Loss of ubiquitination of endogenous GβL promotes RICTOR interaction with mTOR to form mTORC2. IB analysis of WCL or Flag immunoprecipitate derived from GβLKRKR knock-in or wild-type HEK293 cells transfected with Flag–RICTOR construct. Forty-eight hours post-transfection, cells were lysed using CHAPS buffer for Flag immunoprecipitation. The resulting samples were resolved on SDS–PAGE for IB analysis. j, TRAF2 promotes polyubiquitination of GβL, but not the GβL(KRKR) mutant, in cells. IB analysis of WCL or Ni-NTA pull-down products under denaturing conditions derived from HEK293 cells transfected with Flag–TRAF2 and His–Ub, along with HA–GβL, or HA–GβL(KRKR) mutant constructs. k, Compared to ectopic expression of wild-type GβL, reintroducing GβL(KRKR) into GβL-depleted Traf2+/+ cells significantly elevated Akt(pS473). Conversely, in GβL-depleted Traf2−/− cells, reintroducing GβL(KRKR) did not result in any detectable elevation of Akt(pS473). The Traf2−/− MEFs and wild-type MEFs were depleted of endogenous GβL using lentirivus-medicated delivery of short hairpin RNAs (shRNAs) targeting GβL and then reintroduced with GβL and GβL(KRKR) using pBabe-retroviral expression system. The kinase activity of mTORC2/Akt oncogenic signalling in these stable cell lines was analysed by IB analysis of WCL. l, Quantification results for fold differences of GβL binding to Sin1, Rictor and Rptor, or the Akt(pS473) levels, in Gβl−/− MEFs stably expressing GβL or the GβL(KRKR) mutant at the indicated time points of insulin stimulation, as indicated in Fig. 2h. Data from three independent experiments were presented as mean ± s.d. The intensity of each blot generated using ImageJ software were analysed by paired Student’s t-test, *P < 0.05, **P < 0.01. m, GβL polyubiquitination status dictates the dynamic assembly of mTOR complexes in response to EGF stimulation. As such, deficiency in GβL polyubiquitination results in elevated mTORC2 formation and elevated downstream Akt activation. IB analysis of WCL and HA immunoprecipitate derived from Gβl−/− MEFs stably expressing HA–GβL or the ubiquitination-deficient HA–GβL(KRKR) mutant that were generated by retroviral infection, followed by serum starvation for 16 h and exposure to EGF (100 ng ml−1) before harvesting using CHAPS buffer. For uncropped gels, see Supplementary Fig. 1.
Extended Data Figure 5 A schematic model depicting the neutralized regulatory effect of reduced GβL ubiquitination on the total output of mTORC1 signalling in cells.
a, Consistent with a previous study, deletion of Gβl impairs the integrity and kinase activity of mTORC2, but not mTORC1, in mouse embryonic fibroblasts (MEFs) derived from Gβl+/+ and Gβl−/− embryos. Gβl+/+ and Gβl−/− MEFs were transfected with HA-mTOR plasmid, with empty vector as a control. Forty-eight hours post-transfection, cells were lysed using CHAPS buffer for HA immunoprecipitation and IB analysis. b, The schematic illustration of a proposed neutralization model to elucidate the possible underlying mechanism that the observed reduction of mTORC1 abundance resulting from a decrease of GβL ubiquitination in either Traf2−/− cells, GβLKRKR-expressing cells, or at early time points of growth factor stimulation, might be compensated by elevated kinase activity per mTORC1, thus leading to neutralized output of mTORC1 signalling in these cells. This compensation effect is largely derived from reduced GβL ubiquitination-induced elevation of mTORC2 kinase formation that will eventually lead to an increase in AKT-mediated phosphorylation of TSC2, a physiological endogenous inhibitor of mTORC1 signalling that functions largely as a GAP for Rheb. Phosphorylation of TSC2 at multiple sites by AKT releases TSC2 inhibitory effects towards mTORC1, which might further lead to an elevated kinase activity per mTORC1 to balance off its reduction in mTORC1 abundance, thereby leading to minimal perturbation on the total output of mTORC1 signalling in these cells. For uncropped gels, see Supplementary Fig. 1.
Extended Data Figure 6 Deficiency in GβL ubiquitination favours cell survival through elevating the oncogenic mTORC2/AKT signalling.
a, The WD7 motif of GβL is critical for activation of mTORC2 in cells. IB analysis of WCL derived from GβL-depleted OVCAR5 cells stably expressing GβL or indicated GβL mutants. b, c, The SIN1-specific-interacting GβL WD7 motif is critical to maintain mTORC2/AKT signalling to promote cellular survival. GβL-depleted OVCAR5 cells stably expressing WT or various deletion mutants were exposed to indicated concentrations of cisplatin (b) and etoposide (c) for 24 h to measure cell viabilities. Data from three independent experiments were presented as mean ± s.d. and analysed by ANOVA (*P < 0.05, **P < 0.01) d, e, The WD7 motif of GβL is critical for contact-dependent and -independent growth of OVCAR5 cells. Quantification results of colony growth (d) or anchorage independent growth (e) of GβL-depleted OVCAR5 cells stably expressing GβL or indicated GβL mutants. Data were mean ± s.d. from three independent repeats (*P < 0.05, **P < 0.01, ANOVA analysis). f, g, Deficiency in GβL ubiquitination leads to elevated Akt activation in response to insulin stimulation. Gβl−/− MEFs stably expressing HA–GβL and the TRAF2 non-interacting HA–GβL(PEAA) mutant (f) or the ubiquitinaiton-deficient HA–GβL(KRKR) mutant (g) were subjected to serum starvation for 16 h, stimulated with insulin (100 nM) for indicated time points before harvesting for IB analysis of WCL. h, i, Loss of ubiquitination at endogenous GβL promotes resistance to DNA damage drugs. Wild-type or GβLKRKR knock-in HEK293 cells were exposed to indicated concentrations of cisplatin (h) and etoposide (i) for 24 h to measure cell viabilities (biological triplicates, mean ± s.d., *P < 0.05, two-tailed paired Student’s t-test). j, Representative image of the dissected tumours derived from GβL-depleted OVCAR5 cells stably expressing GβL or GβL(KRKR) mutant (upper panel). The weight of individual tumour was shown in the lower panel (n = 5 tumours per group, mean ± s.d.,*P < 0.05, two-tailed paired Student’s t-test). k, Loss of GβL ubiquitination leads to elevated AKT activation in the xenograft tumours. IB analysis of WCL derived from dissected xenografts formed by GβL-depleted OVCAR5 cells stably expressing GβL or GβL(KRKR) mutant. l, The specific AKT kinase inhibitor, MK-2206, inhibits AKT activity in GβL-depleted OVCAR5 cells stably expressing GβL(KRKR) cultured in 10% FBS-containing medium. Cells were treated with MK-2206 at indicated concentration for 2 h and lysed for IB analysis. m–p, Knockdown of endogenous AKT1 or AKT2 inhibits the activation of AKT downstream substrates in GβL-depleted OVCAR5 cells stably expressing GβL(KRKR) (m, n) or in GβLKRKR knock-in HEK293 cells (o, p). q, r, Inhibition of AKT activity by MK-2206 (q) or knocking down AKT1 or AKT2 (r) in GβL-depleted OVCAR5 cells stably expressing GβL(KRKR) promotes cell survival. The cells were incubated with indicated concentrations of cisplatin and etoposide for 24 h. The relative cell viabilities were presented as mean ± s.d. from triple repeats (*P < 0.05, **P < 0.01, ANOVA analysis). s, Knockdown of AKT1 or AKT2 in GβLKRKR knock-in HEK293 cells sensitizes these cells to DNA-damaging drugs. The cell viabilities from triple repeats were presented as mean ± s.d. and analysed by ANOVA (*P < 0.05, **P < 0.01). t, u, AKT activity is critical for contact-dependent and -independent growth of cells expressing the ubiquitination-deficient form of GβL. Quantification of colony growth by GβLKRKR knock-in HEK293 cells (t) or GβL-depleted OVCAR5 cells stably expressing the GβL(KRKR) mutant (u), with or without knocking down AKT1 or AKT2. Data were presented as mean ± s.d. and analysed by ANOVA (*P < 0.05, **P < 0.01). v, Knockdown of AKT1 or AKT2 inhibits anchorage-independent growth of GβL-depleted OVCAR5 cells stably expressing GβL(KRKR). Mean ± s.d. (ANOVA analysis, *P < 0.05, **P < 0.01). For uncroppoed gels, see Supplementary Fig. 1. For tumour data, see Source Data.
Extended Data Figure 7 Cancer-associated ubiquitination-deficient GβL(ΔW297) truncation mutant promotes tumour growth in part via enhancing mTORC2 formation and activating the oncogenic mTORC2/AKT signalling.
a, Compared to wild-type GβL, the cancer-associated GβL(ΔW297) truncation mutant exhibits significantly reduced levels of GβL ubiquitination in cells. Polyubiquitination of GβL in HEK293 cells was examined by transfecting the indicated HA–GβL constructs with His–Ub, followed by IB analysis of WCL and Ni-NTA pull-down products under denaturing conditions. b, Reintroducing the GβL(ΔW297) truncation, compared to GβL, into GβL-depleted A375 cells leads to relatively increased formation of mTORC2 and reduced mTORC1. GβL-depleted A375 cells stably expressing GβL or GβL(ΔW297) were lysed using CHAPS buffer. WCL was filtrated and run through an FPLC Superdex 200 column to collect fractionated cell eluates for subsequent IB analysis. c, Compared to GβL, GβL(ΔW297) is more potent towards activating AKT in cells. IB analysis of WCL derived from Gβl+/+ MEFs or Gβl−/− MEFs transfected with indicated GβL constructs. d, Cancer-associated GβL(ΔW297) truncation enhances mTORC2 activity towards phosphorylating Akt in response to insulin stimulation. Gβl−/− MEFs stably expressing either HA–GβL or the HA–GβL(ΔW297) mutant were serum starved for 16 h, stimulated with insulin (100 nM) for indicated time points, and lysed for IB analysis of WCL. e, Compared to GβL, ectopic expression of the GβL(ΔW297) truncation mutant displays enhanced chemoresistance. GβL-depleted A375 cells stably expressing GβL or GβL(ΔW297) were exposed to indicated concentrations of cisplatin for 24 h. The cell viabilities from triple replicates were presented as mean ± s.d. (*P < 0.05, **P < 0.01, two-tailed paired Student’s t-test). f, Compared to GβL, ectopic expression of the melanoma-associated GβL(ΔW297) truncation is more potent in promoting contact-independent growth of melanoma cells. Representative images of soft agar colony formation by GβL-depleted A375 cells stably expressing GβL or GβL(ΔW297) were shown (triplicate independent experiments, mean ± s.d., **P < 0.01, two-tailed paired Student’s t-test). g, Loss of GβL ubiquitination leads to elevated AKT activation in the xenograft tumours. IB analysis of lysates derived from dissected xenografts formed by GβL-depleted A375 cells stably expressing GβL or the GβL(ΔW297) mutant. h, MK-2206 inhibits AKT activity in GβL-depleted A375 cells stably expressing GβL(ΔW297). Cells were treated with an AKT inhibitor MK-2206 at 1 or 3 μM (with DMSO as vehicle control) for 2 h and lysed for IB analysis. i, Knockdown of AKT1 or AKT2 inhibits the activation of AKT downstream substrates in GβL-depleted A375 cells stably expressing GβL(ΔW297). j, k, Inhibition of AKT activity by MK-2206 (j) or knocking down of AKT1 or AKT2 (k) in GβL-depleted A375 cells expressing GβL(ΔW297) confers sensitivity to DNA-damaging drugs. Cells were incubated with cisplatin or etoposide for 24 h. The relative cell viabilities from triple repeat were presented as mean ± s.d. and analysed by ANOVA (*P < 0.05, **P < 0.01). l, Knockdown of AKT1 or AKT2 inhibits anchorage-independent growth of GβL(ΔW297)-expressing cells A375 cells (three independent experiments, mean ± s.d., ANOVA analysis, *P < 0.05, **P < 0.01). m, A proposed model to describe how the melanoma-associated GβL(ΔW297) truncation mutant disrupts the homeostasis of the two mTOR complexes by impairing polyubiquitination of GβL in cells. Under physiological conditions, the balance of mTORC1 and mTORC2 is tightly controlled by polyubiquitination of GβL on the K305/K313 residues of its WD7 motif. In melanoma cells, the GβL(ΔW297) mutation leads to loss of critical lysine residues for ubiquitination, thus enhancing GβL interaction with SIN1 to promote mTORC2 formation. This subsequently activates the oncogenic AKT signalling to facilitate tumorigenesis. For uncropped gels, see Supplementary Fig. 1.
Extended Data Figure 8 OTUD7B deubiquitinates GβL to promote mTORC2 integrity and signalling activity.
a, Identification of OTUD7B as a specific GβL-interacting deubiquitinase (DUB). IB analysis of WCL and Flag immunoprecipitate derived from HEK293 cells transfected with HA–GβL and Flag-tagged OTU family members of DUBs. b, OTUD7B promotes deubiquitination of GβL in cells in an enzymatic activity-dependent manner. The polyubiquitination status of GβL in HEK293 cells was examined by transfecting HA–GβL and His–Ub with Flag–OTUD7B or activity-deficient OTUD7B(C194A), followed by IB analysis of WCL and Ni-NTA pull-down products under denaturing conditions. c, Knockdown of OTUD7B enhances polyubiquitination of endogenous GβL in cells. IB analysis of WCL or Ni-NTA pull-down products under denaturing conditions derived from control or OTUD7B-depleted HEK293 cells transfected with His–Ub construct. d, K63-linked polyubiquitination of GβL is reduced by ectopic expression of OTUD7B in cells. OTUD7B-depleted HEK293 cells were transfected with CMV-GST-GβL and ubiquitin expression constructs, with or without the Flag–OTUD7B vector. Forty-eight hours post-transfection, cells were lysed using Triton buffer for GST pull-down to elute GST–GβL protein for UB-AQUA-MS analysis of ubiquitin chain linkage in total diGly-purified GβL protein (triplicates, mean ± s.d., **P < 0.01, two-tailed paired Student’s t-test). e, Depletion of Otud7b minimally affects K48-linked polyubiquitination of endogenous GβL. IB analysis of WCL or GβL immunoprecipitate products derived from Otud7b+/+ and Otud7b−/− MEFs transfected with HA–Ub construct. Forty-eight hours post-transfection, the cells were lysed using Triton buffer for GβL immunoprecipitation, with rabbit IgG antibody as negative control. f, Depletion of Otud7b minimally affects the half-life of endogenous GβL protein in cells. IB analysis of WCL derived from Otud7b+/+ and Otud7b−/− MEFs exposed to cycloheximide (CHX, 100 μg ml−1) at indicated time points before harvesting. g, UCH-L1 specifically interacts with RPTOR, but not GβL, in cells. IB analysis of WCL and HA immunoprecipitate derived from HEK293 cells transfected with the GST-UCH-L1 mammalian expression vector, together with HA–RPTOR or HA–GβL plasmids. Cells were lysed using EBC buffer for HA immunoprecipitation procedures. h, UCH-L1 deubiquitinates RPTOR, but not GβL, in cells. IB analysis of WCL or Ni-NTA pull-down products under denaturing conditions derived from HEK293 cells transfected with GST-UCH-L1 and His–Ub constructs, together with HA–RPTOR or HA–GβL plasmids as indicated. i, OTUD7B specifically interacts with GβL, but not RPTOR, in cells. IB analysis of WCL and HA immunoprecipitate derived from HEK293 cells transfected with Flag–OTUD7B, together with HA–RPTOR or HA–GβL constructs where indicated. The cells were lysed using EBC buffer for immunoprecipitation procedures. j, OTUD7B reduces polyubiquitination of GβL, but not RPTOR, in cells. IB analysis of WCL or Ni-NTA pull-down products under denaturing conditions derived from HEK293 cells transfected with Flag–OTUD7B and His–Ub constructs, along with HA–RPTOR or HA–GβL plasmids as indicated. k, Quantification results from three independent experiments showing fold differences of endogenous GβL binding to Sin1, Rictor and Rptor in Otud7b+/+ and Otud7b−/− MEFs, as indicated in Fig. 4c. Data are mean ± s.d. The intensity of each blot generated using ImageJ software was analysed by paired Student’s t-test, *P < 0.05, **P < 0.01. l, Deletion of Otud7b reduces formation of endogenous mTORC2, meanwhile increasing the formation of endogenous mTORC1. Otud7b+/+ and Otud7b−/− MEFs in normal culture medium were lysed using CHAPS buffer to preserve mTOR complex integrity. WCL was fractionated through an FPLC Superdex 200 column. Five hundred microlitres of eluate was collected for each fraction for subsequent SDS–PAGE and IB analysis. m, Deletion of Otud7b does not significantly regulate AKT ubiquitination in cells. IB analysis of WCL or Ni-NTA pull-down products under denaturing conditions derived from Otud7b+/+ and Otud7b−/− MEFs transfected with HA–AKT1 and His–Ub. n, Deletion of Otud7b leads to constant polyubiquitination of GβL that impairs the dynamic fluctuation in the formation of the two mTOR complexes in response to EGF stimulation. IB analysis of WCL, HA immunoprecipitate or Ni-NTA pull-down products derived from Otud7b+/+ and Otud7b−/− MEFs transfected with HA–GβL and His–Ub. Thirty-two hours post-transfection, cells were subjected to serum starvation for 16 h, exposed to EGF (100 ng ml−1), and lysed at indicated time points using CHAPS buffer for HA immunoprecipitation or under denaturing buffer for Ni-NTA pull-down procedures. o, Quantification results from triple replicates showing fold differences of GβL binding to Sin1, Rictor and Rptor, or the Akt(pS473) levels, in Otud7b+/+and Otud7b−/− MEFs stably expressing GβL at the indicated time points of insulin stimulation, as indicated in Fig. 4e and Extended Data Fig. 8p. Data are mean ± s.d. The intensity of each blot generated using ImageJ software was analysed by paired Student’s t-test, *P < 0.05, **P < 0.01. p, q, Loss of Otud7b attenuates mTORC2 kinase activity in response to growth factor stimulation in cells. Otud7b+/+ and Otud7b−/− MEFs were serum starved for 16 h and then treated with insulin (p, 100 nM) or EGF (q, 100 ng ml−1) for indicated time points, and lysed for IB analysis. r, OTUD7B knockdown inhibits mTORC2 kinase activity in cells. IB analysis of WCL derived from OVCAR5 cells stably expressing independent OTUD7B-targeting shRNAs (with shGFP as a negative control). For quantification, AKT(pS473) levels were normalized to total AKT and arbitrarily set at 1.0 in the blot of shGFP cells. s, OTUD7B knockdown sensitizes cells to DNA-damaging drugs. OVCAR5 cells stably expressing independent OTUD7B-targeting lentiviral shRNAs (with shGFP as a negative control) were exposed to etoposide or cisplatin for 24 h to measure cell viabilities. Data from three independent experiments were shown as mean ± s.d. and analysed by ANOVA (*P < 0.05, **P < 0.01). t, u, Knockdown of OTUD7B reduces cell growth and transforming capacity in vitro. Quantification results of colony formation (t) and soft agar colony growth (u) by OVCAR5 cells stably expressing OTUD7B-targeting shRNAs (three independent experiments, mean ± s.d., *P < 0.05, **P < 0.01, ANOVA analysis). For uncropped gels, see Supplementary Fig. 1.
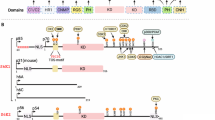
Extended Data Figure 9 Upon physiological stimulation such as growth factor signalling, there is an induced OTUD7B interaction with GβL to enhance mTORC2 activity and oncogenicity primarily through deubiquitinating GβL.
a, b, Deletion of OTUD7B enhances polyubiquitination of GβL largely in cells stimulated with growth factor signalling. IB analysis of WCL and Ni-NTA pull-down products under denaturing conditions derived from HEK293 cells transfected with His–Ub and HA–GβL constructs. Thirty-two hours post-transfection, cells were subjected to serum starvation for 16 h, stimulated with or without EGF (100 ng ml−1) (a) for 16 min, or insulin (100 nM) or 10% FBS-containing medium (b) for 30 min, and lysed at indicated time points. c, OTUD7B interaction with GβL in cells is induced by growth factor stimulation. IB analysis of WCL and HA immunoprecipitate derived from HEK293 cells transfected with HA–GβL construct. Thirty-two hours post-transfection, cells were serum starved for 16 h, then exposed to insulin (100 nM) or EGF (100 ng ml−1), and lysed at indicated time points using EBC buffer for the HA immunoprecipitation procedure. d, Endogenous OTUD7B interaction with GβL is triggered by EGF stimulation. IB analysis of GβL immunoprecipitate or WCL derived from HEK293 cells with or without knockdown of endogenous OTUD7B. Cells were serum starved for 16 h, then stimulated with EGF (100 ng ml−1), and lysed at indicated time points using EBC buffer for immunoprecipitation and IB analysis. See e for quantification results, with levels arbitrarily set to 1.0 at the 0 min time point. e, Quantification results showing fold differences of endogenous GβL binding to OTUD7B in cells at the indicated time points of growth factor stimulation, as indicated in d and Fig. 4f. Data are mean ± s.d. The intensity of each blot from three independent experiments was measured using ImageJ software and analysed by ANOVA, *P < 0.05, **P < 0.01. f, Unlike OTUD7B, UCH-L1 interaction with RPTOR is not regulated by growth stimulation. IB analysis of WCL, GST pull-down products and HA immunoprecipitate derived from HEK293 cells transfected with HA–RPTOR and the GST–UCH-L1 mammalian expression constructs. Thirty-two hours post-transfection, cells were serum starved for 16 h, stimulated with insulin (100 nM) and lysed using EBC buffer at indicated time points for the HA immunoprecipitation procedure. For quantification analysis of endogenous GβL binding to other components, levels were arbitrarily set to 1.0 at the 0 min time point. g, OTUD7B displays a reduced binding affinity to ubiquitination-deficient GβL. IB analysis of WCL and HA immunoprecipitate derived from HEK293 cells transfected with wild-type GβL, GβL(KRKR), or TRAF2-non-interacting GβL(PEAA) constructs. h, A schematic illustration showing different domains of the human OTUD7B protein, as well as various OTUD7B mutants used in this study. i, Deletion of the OTUD7B UBA domain impairs OTUD7B interaction with endogenous GβL. IB analysis of WCL and Flag immunoprecipitate derived from HEK293 cells transfected with OTUD7B or indicated OTUD7B mutant constructs. For quantification results of OTUD7B binding to endogenous GβL levels, levels were arbitrarily set to 1.0 in blot of OTUD7B-expressing cells. j, The UBA domain of OTUD7B is critical for its deubiquitinase activity towards GβL in cells. IB analysis of WCL and Ni-NTA pull-down products under denaturing conditions derived from HEK293 cells transfected with His–Ub and HA–GβL constructs, together with OTUD7B or indicated OTUD7B mutant constructs. k, Deletion of the OTUD7B UBA domain impairs growth-factor-induced OTUD7B interaction with GβL. IB analysis of WCL and Flag immunoprecipitate derived from HEK293 cells transfected with OTUD7B or indicated OTUD7B mutant constructs. Thirty-two hours post-transfection, cells were serum starved for 16 h, stimulated with insulin (100 nM) and lysed using EBC buffer at indicated time points for the immunoprecipitation procedure. l, Deletion of the OTUD7B UBA domain attenuates growth-factor-induced OTUD7B-mediated de-ubiquitination of GβL. IB analysis of WCL and GβL immunoprecipitate products derived from OTUD7B-depleted HEK293 cells stably expressing OTUD7B or indicated OTUD7B mutants. Cells were transfected with His–Ub constructs for 32 h, subjected to serum starvation for 16 h, and then stimulated with or without insulin (100 nM) and lysed using EBC buffer at indicated time points for subsequent IP procedure. m, n, Deletion of the OTUD7B UBA domain attenuates activation of the downstream mTORC2/AKT oncogenic signalling. IB analysis of WCL derived from Otud7b−/− MEFs stably expressing OTUD7B or indicated OTUD7B mutants. The cells were either cultured in 10% FBS-containing culture medium (m), or subjected to serum starvation for 16 h, and then stimulated with or without insulin (n, 100 nM) and lysed using EBC buffer at indicated time points for IB analysis. For quantification, Akt(pS473) levels were normalized to total Akt and arbitrarily set to 1.0 in the first lane. o, OTUD7B promotes deubiquitination of wild-type GβL, but not the GβL(KRKR) mutant, in cells. IB analysis of WCL and Ni-NTA pull-down products under denaturing conditions derived from HEK293 cells transfected with His–Ub and HA–GβL or the ubiquitination-deficient HA–GβL(KRKR) mutant constructs, along with or without Flag–OTUD7B as indicated. p, OTUD7B promotes the integration of GβL, but not GβL(KRKR), into mTORC2 in cells. IB analysis of WCL and HA immunoprecipitate derived from HEK293 cells transfected with HA–GβL or the HA–GβL(KRKR) mutant, together with or without Flag–OTUD7B as indicated. Forty-eight hours post-transfection, cells were lysed using CHAPS buffer for the HA immunoprecipitation procedure to maintain mTOR complex integrity. q, OTUD7B regulates AKT activity primarily through modulating the ubiquitination status of GβL. IB analysis of WCL derived from OTUD7B-depleted or control OVCAR5 cells stably expressing GβL or GβL(KRKR). For quantification of AKT(pS473), levels were normalized to total AKT and arbitrarily set to 1.0 in the first lane. r, OTUD7B governs chemoresistance primarily through modulating the ubiquitination status of GβL. shGFP-infected control or OTUD7B-depleted OVCAR5 cells stably expressing GβL or the GβL(KRKR) mutant were exposed to cisplatin or etoposide for 24 h. Data from three independent experiments were presented as mean ± s.d. and analysed by ANOVA (*P < 0.05, **P < 0.01). s, t, OTUD7B enhances colony growth and anchorage-independent growth of tumour cells primarily through deubiquitinating GβL in cells. Quantitative data of colony growth in plates (s) and in soft agar (t) by control or OTUD7B-depleted OVCAR5 cells stably expressing GβL or the GβL(KRKR) mutant. Representative images of soft agar colonies are shown in t. The result from triple experiments were presented as mean ± s.d. and analysed by ANOVA (**P < 0.01). For uncropped gels, see Supplementary Fig. 1.
Extended Data Figure 10 Downstream of various oncogenic mutations, OTUD7B-mediated activation of mTORC2/AKT signalling plays a physiological role in promoting cancer development.
a, b, Knockdown of endogenous OTUD7B suppresses AKT(pS473) levels in several cancer cell lines in which mTORC2/AKT oncogenic signalling was activated by various upstream oncogenic events. IB analysis of WCL derived from HCT116 (a, PTEN deletion), MCF10A (b, PIK3CA EK mutation), H1650 and H2279 (b, EGFR mutation), and A549 and H157 (b, KRAS mutation) cell lines stably expressing independent OTUD7B-targeting lentiviral shRNAs (with shGFP as a negative control). For quantification results of AKT(pS473) levels, levels were normalized to total AKT and arbitrarily set to 1.0 in the blot of shGFP-expressing cells. c, d, Knockdown of endogenous OTUD7B sensitizes cancer cells to chemotherapeutic drugs. HCT116 (c, PTEN deletion), MCF10A (d, PIK3CA EK mutation), H1650 and H2279 (d, EGFR mutation), and A549 and H157 (d, KRAS mutation) cells stably expressing independent OTUD7B-targeting lentiviral shRNAs were exposed to indicated concentrations of cisplatin or etoposide for 24 h. Cell viability data from triple replicates are presented as mean ± s.d. and were analysed by ANOVA (*P < 0.05, **P < 0.01). e, TCGA DNA sequencing results showing that the OTUD7B gene is amplified at high frequencies in a variety of human cancers, such as breast, lung and pancreatic cancers. f, Kaplan–Meier analysis showing a tight correlation between OTUD7B expression levels and patient survival. Data were from lung cancer patients with low (n = 1,001) and high (n = 925) OTUD7B expression (left panel), and lung adenocarcinoma patients with low (n = 410) and high (n = 310) OTUD7B expression (right panel). Patient number at risk at different times of analyses is indicated at the bottom of the plots. The plots were generated using the KmPlot tool (http://www.kmplot.com/lung). Affymetrix ID 220031_at was used for analyses. g, Lung weight of Otud7b+/+ and Otud7b−/− mice, with (+) or without (−) KrasLA2 transgene expression, at the indicated ages. Data were presented as mean mean ± s.e.m. (16 weeks: Otud7b+/+ mice, n = 6; Otud7b−/− mice, n = 3; 13 weeks: Otud7b+/+KrasLA2 mice, n = 5; Otud7b−/−KrasLA2 mice, n = 5; 16 weeks: Otud7b+/+KrasLA2 mice, n = 8; Otud7b−/−KrasLA2 mice, n = 7; 23 weeks: Otud7b+/+KrasLA2 mice, n = 7; Otud7b−/−KrasLA2 mice, n = 11; **P < 0.01, two-tailed unpaired Student’s t-test). For uncropped gels, see Supplementary Fig. 1. For tumour data, see Source Data for g.
Supplementary information
Supplementary Figure 1
This file contains the original uncropped source images of western blot data for the main Figures and Extended Data Figures. (PDF 36315 kb)
Rights and permissions
About this article
Cite this article
Wang, B., Jie, Z., Joo, D. et al. TRAF2 and OTUD7B govern a ubiquitin-dependent switch that regulates mTORC2 signalling. Nature 545, 365–369 (2017). https://doi.org/10.1038/nature22344
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature22344
This article is cited by
-
Maternal hyperglycemia inhibits pulmonary vasculogenesis during mouse fetal lung development by promoting GβL Ubiquitination-dependent mammalian target of Rapamycin assembly
Diabetology & Metabolic Syndrome (2023)
-
The TRAF2-p62 axis promotes proliferation and survival of liver cancer by activating mTORC1 pathway
Cell Death & Differentiation (2023)
-
Deficiency of BAP1 inhibits neuroblastoma tumorigenesis through destabilization of MYCN
Cell Death & Disease (2023)
-
Deubiquitinases in cancer
Nature Reviews Cancer (2023)
-
Cezanne promoted autophagy through PIK3C3 stabilization and PIK3C2A transcription in lung adenocarcinoma
Cell Death Discovery (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.