Abstract
The heterogeneity of cellular states in cancer has been linked to drug resistance, cancer progression and the presence of cancer cells with properties of normal tissue stem cells1,2. Secreted Wnt signals maintain stem cells in various epithelial tissues, including in lung development and regeneration3,4,5. Here we show that mouse and human lung adenocarcinomas display hierarchical features with two distinct subpopulations, one with high Wnt signalling activity and another forming a niche that provides the Wnt ligand. The Wnt responder cells showed increased tumour propagation ability, suggesting that these cells have features of normal tissue stem cells. Genetic perturbation of Wnt production or signalling suppressed tumour progression. Small-molecule inhibitors targeting essential posttranslational modification of Wnt reduced tumour growth and markedly decreased the proliferative potential of lung cancer cells, leading to improved survival of tumour-bearing mice. These results indicate that strategies for disrupting pathways that maintain stem-like and niche cell phenotypes can translate into effective anti-cancer therapies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Greaves, M. & Maley, C. C. Clonal evolution in cancer. Nature 481, 306–313 (2012)
Kreso, A. & Dick, J. E. Evolution of the cancer stem cell model. Cell Stem Cell 14, 275–291 (2014)
Clevers, H., Loh, K. M. & Nusse, R. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science 346, 1248012 (2014)
Hogan, B. L. et al. Repair and regeneration of the respiratory system: complexity, plasticity, and mechanisms of lung stem cell function. Cell Stem Cell 15, 123–138 (2014)
Herriges, M. & Morrisey, E. E. Lung development: orchestrating the generation and regeneration of a complex organ. Development 141, 502–513 (2014)
de Lau, W., Peng, W. C., Gros, P. & Clevers, H. The R-spondin/Lgr5/Rnf43 module: regulator of Wnt signal strength. Genes Dev. 28, 305–316 (2014)
Schepers, A. G. et al. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science 337, 730–735 (2012)
Huch, M. et al. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J. 32, 2708–2721 (2013)
Boumahdi, S. et al. SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature 511, 246–250 (2014)
Driessens, G., Beck, B., Caauwe, A., Simons, B. D. & Blanpain, C. Defining the mode of tumour growth by clonal analysis. Nature 488, 527–530 (2012)
Meacham, C. E. & Morrison, S. J. Tumour heterogeneity and cancer cell plasticity. Nature 501, 328–337 (2013)
Ilmer, M. et al. RSPO2 enhances canonical Wnt signaling to confer stemness-associated traits to susceptible pancreatic cancer cells. Cancer Res. 75, 1883–1896 (2015)
Zheng, Y. et al. A rare population of CD24+ITGB4+Notchhi cells drives tumor propagation in NSCLC and requires Notch3 for self-renewal. Cancer Cell 24, 59–74 (2013)
Plaks, V., Kong, N. & Werb, Z. The cancer stem cell niche: how essential is the niche in regulating stemness of tumor cells? Cell Stem Cell 16, 225–238 (2015)
Johnson, D. H., Schiller, J. H. & Bunn, P. A. Jr. Recent clinical advances in lung cancer management. J. Clin. Oncol. 32, 973–982 (2014)
Juan, J., Muraguchi, T., Iezza, G., Sears, R. C. & McMahon, M. Diminished WNT → β-catenin → c-MYC signaling is a barrier for malignant progression of BRAFV600E-induced lung tumors. Genes Dev. 28, 561–575 (2014)
Pacheco-Pinedo, E. C. et al. Wnt/β-catenin signaling accelerates mouse lung tumorigenesis by imposing an embryonic distal progenitor phenotype on lung epithelium. J. Clin. Invest. 121, 1935–1945 (2011)
Stewart, D. J. Wnt signaling pathway in non-small cell lung cancer. J. Natl Cancer Inst. 106, djt356 (2014)
Nguyen, D. X. et al. WNT/TCF signaling through LEF1 and HOXB9 mediates lung adenocarcinoma metastasis. Cell 138, 51–62 (2009)
Liu, J. et al. Targeting Wnt-driven cancer through the inhibition of porcupine by LGK974. Proc. Natl Acad. Sci. USA 110, 20224–20229 (2013)
Konermann, S. et al. Genome-scale transcriptional activation by an engineered CRISPR–Cas9 complex. Nature 517, 583–588 (2015)
Willert, J., Epping, M., Pollack, J. R., Brown, P. O. & Nusse, R. A transcriptional response to Wnt protein in human embryonic carcinoma cells. BMC Dev. Biol. 2, 8 (2002)
Rocha, A. S. et al. The angiocrine factor Rspondin3 is a key determinant of liver zonation. Cell Reports 13, 1757–1764 (2015)
Yu, H., Ye, X., Guo, N. & Nathans, J. Frizzled 2 and frizzled 7 function redundantly in convergent extension and closure of the ventricular septum and palate: evidence for a network of interacting genes. Development 139, 4383–4394 (2012)
Gurney, A. et al. Wnt pathway inhibition via the targeting of frizzled receptors results in decreased growth and tumorigenicity of human tumors. Proc. Natl Acad. Sci. USA 109, 11717–11722 (2012)
Chartier, C. et al. Therapeutic targeting of tumor-derived R-Spondin attenuates β-catenin signaling and tumorigenesis in multiple cancer types. Cancer Res. 76, 713–723 (2016)
Feldser, D. M. et al. Stage-specific sensitivity to p53 restoration during lung cancer progression. Nature 468, 572–575 (2010)
Jackson, E. L. et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 15, 3243–3248 (2001)
Marino, S ., Vooijs, M ., van der Gulden, H ., Jonkers, J. & Berns, A. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev. 14, 994–1004 (2000)
Young, N. P., Crowley, D. & Jacks, T. Uncoupling cancer mutations reveals critical timing of p53 loss in sarcomagenesis. Cancer Res. 71, 4040–4047 (2011)
Lee, C. L. et al. Generation of primary tumors with Flp recombinase in FRT-flanked p53 mice. Dis. Model. Mech. 5, 397–402 (2012)
Madisen, L. et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010)
Kuraguchi, M. et al. Adenomatous polyposis coli (APC) is required for normal development of skin and thymus. PLoS Genet. 2, e146 (2006)
Yeddula, N., Xia, Y., Ke, E., Beumer, J. & Verma, I. M. Screening for tumor suppressors: loss of ephrin receptor A2 cooperates with oncogenic KRas in promoting lung adenocarcinoma. Proc. Natl Acad. Sci. USA 112, E6476–E6485 (2015)
Muzumdar, M. D., Tasic, B., Miyamichi, K., Li, L. & Luo, L. A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605 (2007)
Barker, N. et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 (2007)
Sutherland, K. D. et al. Cell of origin of small cell lung cancer: inactivation of Trp53 and Rb1 in distinct cell types of adult mouse lung. Cancer Cell 19, 754–764 (2011)
DuPage, M., Dooley, A. L. & Jacks, T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat. Protoc. 4, 1064–1072 (2009)
Jackson, E. L. et al. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res. 65, 10280–10288 (2005)
Zuber, J. et al. Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nat. Biotechnol. 29, 79–83 (2011)
Korinek, V. et al. Constitutive transcriptional activation by a β-catenin-Tcf complex in APC−/− colon carcinoma. Science 275, 1784–1787 (1997)
Hsu, P. D., Lander, E. S. & Zhang, F. Development and applications of CRISPR–Cas9 for genome engineering. Cell 157, 1262–1278 (2014)
Horlbeck, M. A. et al. Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. eLife 5, e19760 (2016)
Akama-Garren, E. H. et al. A modular assembly platform for rapid generation of DNA constructs. Sci. Rep. 6, 16836 (2016)
Gibson, D. G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009)
Fuerer, C. & Nusse, R. Lentiviral vectors to probe and manipulate the Wnt signaling pathway. PLoS One 5, e9370 (2010)
Hsu, P. D. et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832 (2013)
Ran, F. A. et al. Genome engineering using the CRISPR–Cas9 system. Nat. Protoc. 8, 2281–2308 (2013)
Sánchez-Rivera, F. J. et al. Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature 516, 428–431 (2014)
Vidigal, J. A. & Ventura, A. Rapid and efficient one-step generation of paired gRNA CRISPR–Cas9 libraries. Nat. Commun. 6, 8083 (2015)
Liberzon, A. et al. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015)
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005)
Mootha, V. K. et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003)
Leng, N. et al. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics 29, 1035–1043 (2013)
Abouelhoda, M. I., Kurtz, S. & Ohlebusch, E. Replacing suffix trees with enhanced suffix arrays. J. Discrete Algorithms 2, 53–86 (2004)
Smith, T. F. & Waterman, M. S. Identification of common molecular subsequences. J. Mol. Biol. 147, 195–197 (1981)
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010)
Döring, A., Weese, D., Rausch, T. & Reinert, K. SeqAn an efficient, generic C++ library for sequence analysis. BMC Bioinformatics 9, 11 (2008)
Zhao, M., Lee, W. P., Garrison, E. P. & Marth, G. T. SSW library: an SIMD Smith–Waterman C/C++ library for use in genomic applications. PLoS One 8, e82138 (2013)
Thorvaldsdóttir, H., Robinson, J. T. & Mesirov, J. P. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192 (2013)
Cumming, G. Understanding the New Statistics: Effect sizes, Confidence Intervals, and Meta-Analysis (Routledge, 2012)
Wang, B., Zhao, L., Fish, M., Logan, C. Y. & Nusse, R. Self-renewing diploid Axin2+ cells fuel homeostatic renewal of the liver. Nature 524, 180–185 (2015)
Acknowledgements
We thank D. McFadden and P. Sharp for critical reading of the manuscript and T. Papagiannakopoulos for helpful discussions; H. Clevers for Lgr5CreER/+ mice; Janssen Pharmaceuticals for human tissue; J. Roper for mouse colon adenoma tissue; R. T. Bronson for expertise in animal pathology; Y. Soto-Feliciano and S. Levine for massively parallel sequencing expertise; L. Gilbert, M. Horlbeck and J. Weissman for Lgr5 CRISPR gene activation sgRNA sequences; A. Li for help with generation of TCGA data catalogues; M. Griffin, M. Jennings and G. Paradis for FACS support; E. Vasile for microscopy support; K. Cormier and the Hope Babette Tang (1983) Histology Facility for histology support; S. Bajpay, D. Canner, D. Garcia-Gali, R. Kohn, N. Marjanovich, K. Mercer, J. Replogle and R. Romero for help with experiments; K. Anderson, I. Baptista, A. Deconinck, J. Teixeira and K. Yee for administrative support; and the Swanson Biotechnology Center for excellent core facilities. This work was financially supported by the Transcend Program and Janssen Pharmaceuticals, the Lung Cancer Research Foundation, the Howard Hughes Medical Institute and, in part, by the Cancer Center Support (core) grant P30-CA14051 from the National Cancer Institute. T.T. is supported by the National Cancer Institute (K99 CA187317), the Sigrid Juselius Foundation, the Hope Funds for Cancer Research and the Maud Kuistila Foundation. T.J. is a Howard Hughes Medical Institute Investigator, a David H. Koch Professor of Biology and a Daniel K. Ludwig Scholar.
Author information
Authors and Affiliations
Contributions
T.T. and T.J. designed and directed the study; T.T., K.W., Y.P. and R.A.W. performed all types of experiments reported in the study; F.J.S.-R. performed CRISPR gene activation experiments and analysed CRISPR-mutated loci; N.M.C. and K.H. performed gene expression analysis and N.M.C. performed ISH; N.S.J., L.S. and P.K. performed FACS; R.A. and N.R.K. performed molecular cloning and R.A. quantified Ki67+ nuclei; X.G. performed cell culture experiments; M.C.-B. developed and used microcomputed tomography analysis methodology; W.X. generated shRNA reagents; A.B. conducted bioinformatic analyses; F.J.S.-R., N.S.J., Ö.H.Y., P.K. and A.B. provided conceptual advice; T.T. and T.J. wrote the manuscript with comments from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks H. Clevers and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 Inhibiting Wnt synthesis with the porcupine inhibitor LGK974 suppresses Wnt pathway activation by the R-spondin–Lgr5 axis in primary lung adenocarcinoma cultures.
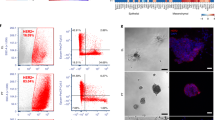
a, Percentage of EdU+ (proliferating) cells in 3D cultures of KPT lung adenocarcinoma (LUAD) cells followed by Wnt pathway stimulation with Rspo1 (1 μg ml−1), Wnt3a (100 ng ml−1) or both (1 μg ml−1 Rspo1 and 100 ng ml−1 Wnt3a), or Wnt pathway inhibition with LGK974 (100 nM) or DKK1 (500 ng ml−1) for seven days (starting at seven days after plating). EdU labelling was performed 16 h before analysis of proliferating cells by flow cytometry. n = 6 wells per condition. b, Percentage of spheroids proliferating in low-density 3D cultures of primary mouse KPT LUAD cells 14 days after plating. n = 8 wells per condition. Representative data from eight replicate experiments; TT5678 and TT5861 identify donor mice. c, qPCR of Porcn transcripts in sublines of a KP LUAD cell line expressing shRNAs targeting Porcn (shPorcn) or control shRNA (shLuc). Quantification of 3D tumour spheroids containing EdU+ cells of KP LUAD cells expressing the indicated shRNAs in response to 100 ng ml−1 Wnt3a or control at six days after plating. Representative data of n = 3 technical replicates, the experiment was performed with three independent cell lines. e, Quantitative real-time PCR (qRT–PCR) analysis of Axin2 and Lgr5 transcripts in 3D cultures of primary KP LUAD cells following Wnt pathway stimulation with Rspo1 (1 μg ml−1), Wnt3a (100 ng ml−1) or both (1 μg ml−1 Rspo1 and 100 ng ml−1 Wnt3a), or Wnt pathway inhibition with LGK974 (100 nM) or DKK1 (1 μg ml−1) for six days (starting at 10 days after plating). Representative data of n = 3 technical replicates. The experiment was carried out three times, each time with cells isolated from a different tumour-bearing mouse. f, Quantification of tumour spheroids containing EdU+ cells per 100 primary KPT LUAD cells 14 days after plating. Combined treatment (1 μg ml−1 Rspo1 and 100 ng ml−1 Wnt3a); LGK974 (100 nM). n = 8 wells per condition. g, Recipient mouse lungs eight weeks after orthotopic transplantation of 30,000 primary tdTomato+ (red) mouse KP LUAD cells. Arrowheads indicate tdTomato+ tumours. Recipient mice were treated with 10 mg per kg per day LGK974 or vehicle for eight weeks, starting from the day of transplantation. Scale bar, 2 mm. The experiment was performed three times, each time with cells isolated from a different (donor) tumour-bearing mouse. h, Wnt-pathway activity measured by detection of firefly luciferase driven by a Wnt-sensitive 7TCF promoter (Topflash assay) in mouse KP LUAD cells stimulated for 24 h with the indicated growth factors. n = 3 technical replicates per condition. Representative data from experiments that were performed with four different cell lines. i–k, qPCR analysis of Rspo2 (i), Rspo3 (j) or Lgr5 (k) transcripts in two independent mouse KP LUAD cell lines (TT5734 and TT5764) expressing SAM components21 driving expression of the indicated genes. l, Flow cytometry analysis of GFP fluorescence in a KP LUAD cell line containing the Lgr5GFP-CreER reporter36 expressing vector control (top) or an sgRNA targeting the transcription start site of Lgr5 (bottom). m, n, qRT–PCR analysis of the Wnt target genes Axin2 (m) and Lgr5 (n) in 3D cultures of sublines of a KP LUAD cell line (TT5764) expressing the CRISPR-activator system driving expression of Rspo2 (Rspo2-a), Rspo3 (Rspo3-a) or Lgr5 (Lgr5-a). Data are mean ± s.d.; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; n.s., not significant; two-way ANOVA (a, b, d, h, m, n) or Student’s two-sided t-test (c, e, f, i–k).
Extended Data Figure 2 Lgr4 and Lgr5 are R-spondin receptors in lung adenocarcinoma.
a, qRT–PCR analysis of Lgr4, Lgr5 and Lgr6 transcripts in sublines of a KP LUAD cell line stably expressing shLgr4, shLgr5, shLgr6 or shLuciferase (shLuc) control. Note minimal effects of the indicated shRNAs on other Lgr5 family members. n = 3 technical replicates per condition; representative data from three experiments carried out in different cell lines. b–e, Formation of 3D tumour spheroids of sublines of a KP LUAD cell line expressing the indicated shRNAs in response to 1 μg ml−1 Rspo1 (b, c) or 100 ng ml−1 Wnt3a (d, e) six days after plating. n = 8 wells per condition, representative data from three experiments carried out in different cell lines. f, g, No difference in cell morphology (f) or growth rate (g) in sublines of a KP LUAD cell line expressing shLgr4, shLgr5, shLgr6 or control shLuciferase (shLuc) over six days in two-dimensional cell culture. Scale bars, 500 μm (b, d) and 200 μm (f). Data are mean + s.d.; *P < 0.05; **P < 0.01; ***P < 0.001; Student’s two-sided t-test (a) or two-way ANOVA (c, e, g). All experiments were performed three times, each time with a different KP LUAD cell line.
Extended Data Figure 3 Wnt ligands predominantly produced by cancer cells drive activation of the Wnt signalling pathway in lung adenocarcinoma.
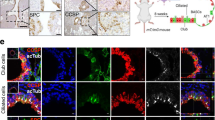
a, Schematic representation of the lentiviral vector46 used to transduce a KPT LUAD cell line, followed by puromycin selection. b, tdTomato and 7TCF::Luciferase signals at baseline (0 h) and 48 h after treatment with 10 mg per kg per day LGK974 or vehicle. Red arrows indicate two subcutaneous tumours with reduced 7TCF-dependent bioluminescence in response to 48 h of LGK974 treatment. c, Suppression of 7TCF-driven bioluminescence by LGK974 relative to the tdTomato signal in mice with subcutaneous transplants of the KPT LUAD cell line stably expressing the 7TCF::Luciferase-PGK-Puro lentivirus. n = 6 tumours, three mice per group; representative data from two independent experiments. Data are mean ± s.d.; Student’s two-sided t-test. d, Haematoxylin and eosin (HE) or β-catenin staining in KP adenomas or in adenocarcinomas. e, Immunofluorescence for β-catenin (red) and porcupine (green) in a KP LUAD. f, ISH for Axin2 mRNA (purple, arrowheads) in KP lung adenomas or adenocarcinomas. g, Immunofluorescence for porcupine (green, white arrowheads) and ISH for Axin2 (red, yellow arrowheads) in a KP LUAD. h, Immunofluorescence for porcupine (green) in an autochthonous grade 3 KPT adenocarcinoma. Arrowheads indicate peritumoural porcupine+ cells, which are tdTomato− (that is, not cancer cells). i, CD11b (green) and porcupine (red) immunofluorescence in a peritumoural region. White line delineates the tumour (T). j, Immunohistochemistry for β-catenin or porcupine in human LUAD. Arrowheads indicate cells with nuclear β-catenin. 65 human LUAD tumours in two tissue microarrays were analysed. Scale bars, 100 μm (d, e, h–j), 50 μm (f) and 10 μm (g). k, Comparison of PORCN gene expression in tumours versus normal tissue in the TCGA lung adenocarcinoma cohort: Empirical cumulative density function (CDF) plots of standardized gene expression values are shown. A right-shift indicates relatively higher expression, with P values indicated to assess statistical significance (Kolmogorov–Smirnov test).
Extended Data Figure 4 Expression of porcupine in lung adenocarcinoma, the normal lung and in stem cell niches of the intestine and liver.
a, ISH for Porcn in a grade 3 KP lung adenocarcinoma. b, c, Immunohistochemistry for porcupine (brown) (b), or Porcn ISH in the normal lung (c). Arrowheads indicate porcupine expression in bronchioles. d, Immunohistochemistry for β-catenin (brown) and porcupine (purple) in the small intestine. Note porcupine expression in intestinal crypts (black arrowheads), the location of Lgr5+ intestinal stem cells36, as well as in transit-amplifying cells (blue arrowheads) and stromal cells (dark green). e, Immunostaining for porcupine (brown) and glutamine synthetase (GS, purple) expression localizes to areas around the central vein of the liver (e), coinciding with the location of Lgr5+Axin2+ liver stem cells8,62. Scale bars, 100 μm.
Extended Data Figure 5 Analysis of the Porcn locus following CRISPR–Cas9-mediated genome editing in vivo.
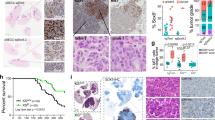
a, Haematoxylin and eosin staining of KP LUAD-bearing lungs generated with pSECC–sgTom or pSECC–sgPorcn and quantification of the proportion of adenomas versus adenocarcinomas at 12 weeks after tumour initiation. Scale bar, 2 mm. Data are mean ± s.d.; Student’s two-sided t-test; n = 5 mice per group. b, Massively parallel sequencing analysis of allelic fractions of the Porcn locus in lung lobes containing microscopic tumours (sgPorcn.2 L) or microdissected macroscopic tumours (sgPorcn.2 T) induced in KP mice using pSECC–sgPorcn.2, or in lung lobes or macroscopic tumours induced in KP mice using pSECC–sgTom.2 (sgTom.2 T/L). FS, frameshift mutation; low freq, low-frequency mutation event; NFS, non-frameshift mutation; WT, wild-type read. Note predominantly wild-type or non-frameshift reads in microdissected tumours, whereas mutations in tumours containing microscopic tumours have introduced frameshifts. The large contribution of wild-type reads in sgPorcn.2 L samples is owing to domination of the normal stroma in whole-lobe samples, whereas wild-type reads in sgPorcn.2 T indicate cancer cells where genome editing did not function, as in whole-tumour samples, tumour cells are expected to contribute at least 50% (ref. 49). c, Qualitative analysis of mutations introduced by sgPorcn.2 in vivo. bp, base pair (indicates the size of the insertion or deletion); INS, insertion; DEL, deletion. Ratio indicates frequency of event across the 15 samples analysed.
Extended Data Figure 6 Lgr5 and Lgr4 are expressed in lung adenocarcinoma and Lgr5 marks cells with increased tumour-forming ability.
a, qPCR analysis of Lgr5 gene expression in KP LUAD tumours microdissected at 9 weeks (adenomas) or 20 weeks (adenocarcinomas) after initiation with adenoviral Cre. n = 6 tumours per group. b, ISH for Lgr5 or Lgr4 mRNA (purple) in grade 3 KP LUAD adenocarcinomas 12 weeks after tumour induction with AdCre. c, d, FACS sorting (d) of Lgr5+ (GFP+) cells in a cultured KP LUAD cell line containing the Lgr5GFP-CreER/+ reporter allele (KP;Lgr5GFP-CreER/+), followed by qRT–PCR analysis (c) of Lgr4 expression in Lgr5+ cells in two independent cell lines (TT1937 and TT6280). This experiment was performed once. e, FACS of GFP+ and GFP− cells isolated from KPT;Lgr5GFP-CreER/+ primary LUAD 14 weeks after tumour initiation with intratracheally administered AdCre. The FACS plot is gated on tdTomato+CD11b−CD31−CD45−TER119− cells. Note bleeding of the tdTomato signal to the GFP channel in the panel on the right. Gates were drawn as shown to increase cell yield at the cost of purity to enrich for Lgr5+ cells. Such FACS sorting was performed on 21 KPT;Lgr5GFP-CreER/+ mice. f, Recipient mouse lungs, 12 weeks after orthotopic transplantation of 15,000 primary Lgr5+tdTomato+ or Lgr5−tdTomato+ primary mouse LUAD cells. Arrowheads indicate tdTomato+ tumours (red). Representative data from three replicate experiments. g, Quantification of tumours per 1,000 cells in recipient mouse lungs 12 weeks after orthotopic transplantation of 15,000 primary Lgr5+/tdTomato+ or Lgr5−/tdTomato+ primary mouse LUAD cells. n = 3 recipient mice per group; representative data from three replicate experiments. h, Number of membrane-associated GFP+ (mGFP+) clones following lineage-tracing in established subcutaneous KP;Lgr5CreER/+;Rosa26LSL-mTmG/+ LUAD primary transplants (see Fig. 3c). n = 9 tumours/time point. Scale bars, 2 mm (f) and 100 μm (b). Data are mean ± s.d.; *P < 0.05; n.s., not significant; Student’s two-sided t-test (a, c, g, h).
Extended Data Figure 7 Phenotypic plasticity of Lgr5+ cells that reside in porcupine+ niches in lung, pancreatic and colon tumours.
a, Immunofluorescence for GFP (green) and EpCAM (red) in a subcutaneous transplant established from a single-cell clone of a KPT;Lgr5CreER/+ cell line. b, Immunofluorescence for GFP (green) and porcupine (red) in a subcutaneous transplant established from a single-cell clone of a KPT;Lgr5CreER/+ cell line. a, b, Data are representative of four replicate experiments, each with a different KP;Lgr5CreER/+ cell line. c, ISH for Lgr5 mRNA (purple) in KP;Pdx1::Cre PDAC. Representative of three PDAC tumours analysed. d, Immunofluorescence staining for GFP (green) in a tdTomato+ (red) autochthonous KP;Lgr5GFP-CreER/+;Rosa26tdTomato/+;Pdx1::Cre PDAC. e, Quantification of primary spheroids containing EdU+ cells per 100 Lgr5+tdTomato+ or Lgr5−tdTomato+ primary mouse PDAC cells plated. n = 4 wells per group. Data are mean ± s.d.; *P < 0.05; Student’s two-sided t-test. f, Immunofluorescence staining for GFP (green) and porcupine (red) in autochthonous KP;Lgr5GFP-CreER/+;Pdx1::Cre PDAC. Note juxtaposition of Lgr5+ and porcupine+ cells in the tumours. d, f, Representative data from six KP;Lgr5GFP-CreER/+;Pdx1::Cre PDAC tumours analysed. g, Immunofluorescence staining for GFP (green) and porcupine in an autochthonous ApcΔ/Δ;Lgr5GFP-CreER/+intestinal adenoma. Again, note juxtaposition of Lgr5+ and porcupine+ cells in the tumours. n = 3 tumour samples. h, Immunohistochemistry for porcupine (brown) in human colorectal adenocarcinoma. Five human colorectal adenocarcinoma samples were analysed. Scale bars, 1 mm (a), 100 μm (b, c, d, f and h (top), g) and 10 μm (c, d, f and h (bottom)).
Extended Data Figure 8 Wnt pathway activation correlates with poor survival in human lung adenocarcinoma, pancreatic ductal adenocarcinoma and mesothelioma, but not in human squamous cell lung cancer; analysis of the Lgr4 and Lgr5 loci following CRISPR–Cas9-mediated genome editing in vivo.
a, Kaplan–Meier survival curve comparing the 20% strongest (red, n = 91) and weakest (blue, n = 92) correlations of the Wnt signature from ref. 22 and patient samples from the TCGA lung adenocarcinoma cohort. b, Empirical CDF plots of standardized gene expression values showing a correlation between the Wnt pathway activation gene expression signature correlation score and histological grade of primary tumours. A right-shift indicates relatively higher expression, with P values indicated to assess statistical significance (Kolmogorov–Smirnov test). c–e, Kaplan–Meier survival curves comparing the 20% strongest (red) and weakest (blue) correlations of the Wnt signature from ref. 22 and patients samples from the TCGA squamous cell lung cancer (c, most-correlated (red) n = 100, least-correlated (blue) n = 100), pancreatic ductal adenocarcinoma (d, most-correlated (red) n = 34, least-correlated (blue) n = 34) and mesothelioma (e, most-correlated (red) n = 17, least-correlated (blue) n = 18) cohorts. f, Massively parallel sequencing analysis of allelic fractions of the Lgr4 and Lgr5 loci in lung lobes containing microscopic tumours (lobe) or microdissected macroscopic tumours (tumour) induced in KrasLSL-G12D/+;Trp53flox/flox;Rosa26LSL-Cas9+2a+eGFP/+ mice using hU6::sgLgr4-sU6::sgLgr5-EFS::Cre (pU2SEC) or hU6::sgTom-EFS::Cre (pUSEC) lentiviral vectors. FS, frameshift mutation; NFS, non-frameshift mutation; WT, wild-type read. Note predominantly wild-type or non-frameshift reads in microdissected tumours, whereas mutations in tumours containing microscopic tumours have introduced frameshifts. The large contribution of wild-type reads in lobe samples is owing to domination of the normal stroma in whole-lobe samples, whereas wild-type reads in Lgr4/Lgr5 co-targeted tumours indicate cancer cells where genome editing did not function, as in whole tumour samples, tumour cells are expected to contribute at least 50% (ref. 49). g, h, Qualitative analysis of mutations introduced by sgLgr4 or sgLgr5 in vivo. bp, base pair (indicates size of insertion/deletion); INS, insertion; DEL, deletion. Ratio indicates frequency of event across all samples analysed. P values are indicated (log-rank test).
Extended Data Figure 9 Characterization of the niche for stem-like cells in lung adenocarcinoma.
a, qPCR analysis of Rspo gene expression in 16 KP LUAD tumours, normalized to Actb expression. Tumours were collected at 16 weeks after initiation with adenoviral Cre. ISH for Rspo1 mRNA (purple, arrowheads) in a KP LUAD tumour. Note the Rspo1 transcripts in endothelial cells. b, qPCR for Rspo1 and Rspo3 in tdTomato+ tumour cells (tumour), CD31+ endothelial cells and the rest of the cells (stroma) in microdissected KPT LUAD tumours following sorting. The expression of Pecam1 (which encodes CD31) was found to be >400-fold enriched in the CD31+ fraction compared to the stroma (not shown). n = 3 mice, representative of two replicate experiments. c, Heatmap showing relative expression levels of Porcn and the 19 mouse Wnt genes on the basis of the qPCR analysis in sorted tdTomato+ KP LUAD cells (T) versus tdTomato− stromal cells (S) in microdissected tumours collected at 20 weeks after tumour initiation (a time point when most tumours are adenocarcinomas). d, Volcano plot of qPCR array gene expression analysis showing statistically significant differentially expressed genes (in red, Fzd receptors are circled). x axis is the log2 fold change (tumour/stroma) and y axis is the –log10P value of the differential enrichment (two-sided t-test). e, qPCR analysis of Wnt5a, Wnt7a and Wnt7b gene expression in KP tumours microdissected at 9 weeks (adenomas) or 20 weeks (adenocarcinomas) after initiation with adenoviral Cre. n = 6 mice, representative of two replicate experiments. f, Comparison of WNT gene expression in tumours versus normal tissue in the TCGA lung adenocarcinoma cohort: Empirical CDF plots of standardized gene expression values for WNT5A and WNT7B are shown. A right-shift indicates relatively higher expression, with P values indicated to assess statistical significance (Kolmogorov–Smirnov test). g, Heatmap showing relative expression levels of Lrp5, Lrp6 and nine mouse Fzd genes on the basis of the qPCR analysis in sorted tdTomato+ KP LUAD cells (T) versus tdTomato− stromal cells (S) in microdissected tumours collected at 20 weeks after tumour initiation (a time point when most tumours are adenocarcinomas). h, qPCR analysis of eight Fzd receptors in KP tumours microdissected at 9 weeks (adenomas) or 20 weeks (adenocarcinomas) after initiation with adenoviral Cre. n = 3 mice. Data are mean ± s.d.; *P < 0.05; Student’s two-sided t-test (b, e, h).
Extended Data Figure 10 Porcupine inhibition suppresses Wnt pathway activity, progression and proliferative potential in autochthonous mouse KP lung adenocarcinomas.
a, qPCR analysis of Axin2 and Lgr5 transcripts in KP LUAD tumours two weeks after treatment with 10 mg per kg per day LGK974 or vehicle. Treatment was started at 11 weeks after tumour initiation. n = 6 tumours per group. b, Quantification of μCT data showing change in tumour volume compared to baseline (obtained at 76 days after tumour initiation, dashed line) after four weeks of 10 mg per kg per day LGK974 or vehicle control. c, Recipient mouse lungs four weeks after orthotopic GEMM-DA of 50,000 primary tdTomato+ (red) mouse LUAD cells. Arrowheads indicate tdTomato+ tumours. Donor mice bearing autochthonous KPT LUAD tumours were treated for two weeks with LGK974 or vehicle (starting at 84 days after tumour induction). The recipient mice were treated with LGK974 or vehicle for four weeks. d, tdTomato+ tumours in sections from lungs in c containing EdU+ cells (white arrowheads) or not containing EdU+ cells (yellow arrowheads). Scale bars, 2 mm (c), 500 μm (d (top)) and 100 μm (d (bottom)). e, Quantification of EdU+ (black) or EdU− (grey) tumours per section through the lungs depicted in c, d. n = 5 (vehicle–vehicle), representative data from three replicate experiments. Data are mean ± s.d.; *P < 0.05; Student’s two-sided t-test (a, b) or two-way ANOVA (e).
Supplementary information
Supplementary Information
This file contains additional discussion on the differences between R-spondin and Wnt ligand stimulation. (PDF 51 kb)
Supplementary Tables
This file contains Supplementary Tables 1-4. (XLSX 27 kb)
Source data
Rights and permissions
About this article
Cite this article
Tammela, T., Sanchez-Rivera, F., Cetinbas, N. et al. A Wnt-producing niche drives proliferative potential and progression in lung adenocarcinoma. Nature 545, 355–359 (2017). https://doi.org/10.1038/nature22334
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature22334
This article is cited by
-
Lung endothelium exploits susceptible tumor cell states to instruct metastatic latency
Nature Cancer (2024)
-
WNT/β-catenin regulatory roles on PD-(L)1 and immunotherapy responses
Clinical and Experimental Medicine (2024)
-
Cancer cell plasticity during tumor progression, metastasis and response to therapy
Nature Cancer (2023)
-
Tumor cell plasticity in targeted therapy-induced resistance: mechanisms and new strategies
Signal Transduction and Targeted Therapy (2023)
-
Oxidative stress-triggered Wnt signaling perturbation characterizes the tipping point of lung adeno-to-squamous transdifferentiation
Signal Transduction and Targeted Therapy (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.