Abstract
The Notch signalling pathway mediates cell fate decisions1,2 and is tumour suppressive or oncogenic depending on the context2,3. During lung development, Notch pathway activation inhibits the differentiation of precursor cells to a neuroendocrine fate4,5,6. In small-cell lung cancer, an aggressive neuroendocrine lung cancer7, loss-of-function mutations in NOTCH genes and the inhibitory effects of ectopic Notch activation indicate that Notch signalling is tumour suppressive8,9. Here we show that Notch signalling can be both tumour suppressive and pro-tumorigenic in small-cell lung cancer. Endogenous activation of the Notch pathway results in a neuroendocrine to non-neuroendocrine fate switch in 10–50% of tumour cells in a mouse model of small-cell lung cancer and in human tumours. This switch is mediated in part by Rest (also known as Nrsf), a transcriptional repressor that inhibits neuroendocrine gene expression. Non-neuroendocrine Notch-active small-cell lung cancer cells are slow growing, consistent with a tumour-suppressive role for Notch, but these cells are also relatively chemoresistant and provide trophic support to neuroendocrine tumour cells, consistent with a pro-tumorigenic role. Importantly, Notch blockade in combination with chemotherapy suppresses tumour growth and delays relapse in pre-clinical models. Thus, small-cell lung cancer tumours generate their own microenvironment via activation of Notch signalling in a subset of tumour cells, and the presence of these cells may serve as a biomarker for the use of Notch pathway inhibitors in combination with chemotherapy in select patients with small-cell lung cancer.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Hori, K., Sen, A. & Artavanis-Tsakonas, S. Notch signaling at a glance. J. Cell Sci. 126, 2135–2140 (2013)
Ntziachristos, P., Lim, J. S., Sage, J. & Aifantis, I. From fly wings to targeted cancer therapies: a centennial for notch signaling. Cancer Cell 25, 318–334 (2014)
Koch, U. & Radtke, F. Notch and cancer: a double-edged sword. Cell. Mol. Life Sci. 64, 2746–2762 (2007)
Ito, T. et al. Basic helix-loop-helix transcription factors regulate the neuroendocrine differentiation of fetal mouse pulmonary epithelium. Development 127, 3913–3921 (2000)
Morimoto, M., Nishinakamura, R., Saga, Y. & Kopan, R. Different assemblies of Notch receptors coordinate the distribution of the major bronchial Clara, ciliated and neuroendocrine cells. Development 139, 4365–4373 (2012)
Tsao, P. N. et al. Notch signaling controls the balance of ciliated and secretory cell fates in developing airways. Development 136, 2297–2307 (2009)
Pietanza, M. C., Byers, L. A., Minna, J. D. & Rudin, C. M. Small cell lung cancer: will recent progress lead to improved outcomes? Clin. Cancer Res. 21, 2244–2255 (2015)
George, J. et al. Comprehensive genomic profiles of small cell lung cancer. Nature 524, 47–53 (2015)
Sriuranpong, V. et al. Notch signaling induces cell cycle arrest in small cell lung cancer cells. Cancer Res. 61, 3200–3205 (2001)
Schaffer, B. E. et al. Loss of p130 accelerates tumor development in a mouse model for human small-cell lung carcinoma. Cancer Res. 70, 3877–3883 (2010)
Fre, S. et al. Notch lineages and activity in intestinal stem cells determined by a new set of knock-in mice. PLoS ONE 6, e25785 (2011)
Krebs, L. T., Deftos, M. L., Bevan, M. J. & Gridley, T. The Nrarp gene encodes an ankyrin-repeat protein that is transcriptionally regulated by the notch signaling pathway. Dev. Biol. 238, 110–119 (2001)
Borges, M. et al. An achaete-scute homologue essential for neuroendocrine differentiation in the lung. Nature 386, 852–855 (1997)
Jiang, T. et al. Achaete-scute complex homologue 1 regulates tumor-initiating capacity in human small cell lung cancer. Cancer Res. 69, 845–854 (2009)
Kaur, G. et al. Bromodomain and hedgehog pathway targets in small cell lung cancer. Cancer Lett. 371, 225–239 (2016)
Milano, J. et al. Modulation of notch processing by gamma-secretase inhibitors causes intestinal goblet cell metaplasia and induction of genes known to specify gut secretory lineage differentiation. Toxicol. Sci. 82, 341–358 (2004)
Calbo, J. et al. A functional role for tumor cell heterogeneity in a mouse model of small cell lung cancer. Cancer Cell 19, 244–256 (2011)
Sutherland, K. D. et al. Cell of origin of small cell lung cancer: inactivation of Trp53 and rb1 in distinct cell types of adult mouse lung. Cancer Cell 19, 754–764 (2011)
Luca, V. C. et al. Structural basis for Notch1 engagement of Delta-like 4. Science 347, 847–853 (2015)
Yen, W. C. et al. Targeting Notch signaling with a Notch2/Notch3 antagonist (tarextumab) inhibits tumor growth and decreases tumor-initiating cell frequency. Clin. Cancer Res. 21, 2084–2095 (2015)
Morrison, S. J. et al. Transient Notch activation initiates an irreversible switch from neurogenesis to gliogenesis by neural crest stem cells. Cell 101, 499–510 (2000)
Ball, D. W. Achaete-scute homolog-1 and Notch in lung neuroendocrine development and cancer. Cancer Lett. 204, 159–169 (2004)
Mori, N., Schoenherr, C., Vandenbergh, D. J. & Anderson, D. J. A common silencer element in the SCG10 and type II Na+ channel genes binds a factor present in nonneuronal cells but not in neuronal cells. Neuron 9, 45–54 (1992)
Chong, J. A. et al. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell 80, 949–957 (1995)
Ballas, N. & Mandel, G. The many faces of REST oversee epigenetic programming of neuronal genes. Curr. Opin. Neurobiol. 15, 500–506 (2005)
Pietras, K. & Ostman, A. Hallmarks of cancer: interactions with the tumor stroma. Exp. Cell Res. 316, 1324–1331 (2010)
Banker, G. A. Trophic interactions between astroglial cells and hippocampal neurons in culture. Science 209, 809–810 (1980)
Kadomatsu, K. & Muramatsu, T. Midkine and pleiotrophin in neural development and cancer. Cancer Lett. 204, 127–143 (2004)
Williamson, S. C. et al. Vasculogenic mimicry in small cell lung cancer. Nature Commun. 7, 13322 (2016)
Madisen, L. et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nature Neurosci. 13, 133–140 (2010)
Cheung, A. F., Dupage, M. J., Dong, H. K., Chen, J. & Jacks, T. Regulated expression of a tumor-associated antigen reveals multiple levels of T-cell tolerance in a mouse model of lung cancer. Cancer Res. 68, 9459–9468 (2008)
Jahchan, N. S. et al. A drug repositioning approach identifies tricyclic antidepressants as inhibitors of small cell lung cancer and other neuroendocrine tumors. Cancer Discov. 3, 1364–1377 (2013)
Demitrack, E. S. et al. Notch signaling regulates gastric antral LGR5 stem cell function. EMBO J. 34, 2522–2536 (2015)
Rothe, C. et al. The human combinatorial antibody library HuCAL GOLD combines diversification of all six CDRs according to the natural immune system with a novel display method for efficient selection of high-affinity antibodies. J. Mol. Biol. 376, 1182–1200 (2008)
Brahmer, J. R. & Ettinger, D. S. Carboplatin in the treatment of small cell lung cancer. Oncologist 3, 143–154 (1998)
Shi, Y. et al. Cisplatin combined with irinotecan or etoposide for untreated extensive-stage small cell lung cancer: a multicenter randomized controlled clinical trial. Thorac. Cancer 6, 785–791 (2015)
Wu, Y. et al. Therapeutic antibody targeting of individual Notch receptors. Nature 464, 1052–1057 (2010)
Riccio, O. et al. Loss of intestinal crypt progenitor cells owing to inactivation of both Notch1 and Notch2 is accompanied by derepression of CDK inhibitors p27Kip1 and p57Kip2. EMBO Rep. 9, 377–383 (2008)
Jahchan, N. S. et al. Identification and targeting of long-term tumor-propagating cells in small cell lung cancer. Cell Reports 16, 644–656 (2016)
Nadeau, H. & Lester, H. A. NRSF causes cAMP-sensitive suppression of sodium current in cultured hippocampal neurons. J. Neurophysiol. 88, 409–421 (2002)
Sanjana, N. E., Shalem, O. & Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nature Methods 11, 783–784 (2014)
Park, K. S. et al. A crucial requirement for Hedgehog signaling in small cell lung cancer. Nature Med. 17, 1504–1508 (2011)
Zheng, Y. et al. A rare population of CD24+ITGB4+Notchhi cells drives tumor propagation in NSCLC and requires Notch3 for self-renewal. Cancer Cell 24, 59–74 (2013)
Tuominen, V. J., Ruotoistenmäki, S., Viitanen, A., Jumppanen, M. & Isola, J. ImmunoRatio: a publicly available web application for quantitative image analysis of estrogen receptor (ER), progesterone receptor (PR), and Ki-67. Breast Cancer Res. 12, R56 (2010)
Mootha, V. K. et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature Genet. 34, 267–273 (2003)
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005)
Kuleshov, M. V. et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44 (W1), W90–W97 (2016)
Chen, E. Y. et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 (2013)
Hodges, P. E. et al. Annotating the human proteome: the Human Proteome Survey Database (HumanPSD) and an in-depth target database for G protein-coupled receptors (GPCR-PD) from Incyte Genomics. Nucleic Acids Res. 30, 137–141 (2002)
Matys, V. et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 34, D108–D110 (2006)
Denny, S. K. et al. Nfib promotes metastasis through a widespread increase in chromatin accessibility. Cell 166, 328–342 (2016)
Yang, J. S. et al. OASIS: online application for the survival analysis of lifespan assays performed in aging research. PLoS ONE 6, e23525 (2011)
Corish, P. & Tyler-Smith, C. Attenuation of green fluorescent protein half-life in mammalian cells. Protein Eng. 12, 1035–1040 (1999)
Hirata, H. et al. Oscillatory expression of the bHLH factor Hes1 regulated by a negative feedback loop. Science 298, 840–843 (2002)
Kobayashi, T. et al. The cyclic gene Hes1 contributes to diverse differentiation responses of embryonic stem cells. Genes Dev. 23, 1870–1875 (2009)
Acknowledgements
We thank A. Berns for Trp53lox mice, S. Fre and S. Artavanis-Tsakonas for Hes1GFP mice, M. Winslow, M. Krasnow, T. Rando, A. Sweet-Cordero, Y. Ouadah and Sage laboratory members for suggestions on the manuscript, and P. Lovelace and J. Ho (fluorescence-activated cell sorting (FACS)), P. Chu (histology), S. Sim and the Stanford Protein and Nucleic Acid (PAN) facility for technical support. We thank the Tumorothèque des Hôpitaux Universitaires de l’Est Parisien (HUEP; the East Paris University Hospitals Tumor Bio-bank) for SCLC samples and N. Rabbe-Mathiot for immunostaining support. This work was supported by the Howard Hughes Medical Institute (K.C.G.), the National Institutes of Health (J.S. R01 CA201513), A*STAR Singapore (J.S.L.), Soutien à la Recherche Clinique (Fondation du Souffle and Fonds de Recherche en Santé Respiratoire, M.W.), and the Legs Poix (Chancellerie des Universités de Paris, M.W.). J.S. is the Harriet and Mary Zelencik Scientist in Children’s Cancer and Blood Diseases.
Author information
Authors and Affiliations
Contributions
J.S.L., M.W., K.C.G., T.H., C.L.M., and J.S. conceptualized the study. J.S.L. designed and performed cell culture assays and experiments using the mouse model. A.I. contributed to immunoblotting and immunostaining. M.M.F., B.C., G.O., J.C., A.M.K., and C.L.M. performed tarextumab and chemotherapy treatments and analysis. S.C. generated TKO allografts. V.C.L. and K.C.G. provided recombinant Dll4. D.Y. and N.S.J. contributed reagents and to data interpretation. C.H., M.A., M.W., and C.K. performed and analysed HES1 immunostaining on patient samples. Y.-W.L. contributed to Luminex assays. J.S.L., T.H., C.L.M., and J.S. wrote the manuscript with input from all authors. J.S. supervised the study, data interpretation, and manuscript preparation.
Corresponding author
Ethics declarations
Competing interests
M.M.F., B.C., G.O., J.C., Y.-W.L., A.M.K., T.H., and C.L.M. are employees and stockholders of OncoMed Pharmaceuticals.
Additional information
Reviewer Information Nature thanks A. Berns, L. Miele and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
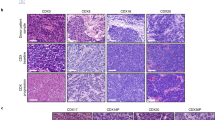
Extended Data Figure 1 SCLC tumours contain a population of Hes1-positive tumour cells.
a, Quantification of the frequency (freq.) of Hes1pos cells in TKO tumours at different stages of tumour development. Scores: 0 (0%), 1 (1–20%), 2 (20–60%), 3 (>60%). Hyperplasias (n = 26, from five mice, defined by area less than 16,600 μm2) and early tumours (n = 69, from five mice) were analysed 3 months after Ad-CMV-Cre; late tumours (n = 83, from five mice) and liver metastases (n = 54, from five mice) were analysed 6–7 months after Ad-CMV-Cre (data for the late tumours are shown for comparison and are the same as in Fig. 1b). b, As in a, for human SCLC tumour microarray sections and segregated by clinical stage. H scores: stage I (123.9; n = 71 sections), stage II (135.3; n = 68 sections), stages III + IV (148.5; n = 33 sections). c, Representative immunofluorescence for GFP in TKO Hes1GFP/+ SCLC tumours. d, Representative flow cytometry plots of cells isolated from pooled tumours from a TKO Hes1GFP/+ mouse. Arrows depict the sequential gating strategy for enriching for single, live (by exclusion of 7-aminoactinomycin D (7-AAD)), and lineage (CD45, CD31, TER-119)-negative cells. CD24 labels more than 98% of Cre-recombined cells and thus further enriches for tumour cells39. e, Immunofluorescence of a TKO Rosa26lox-stop-lox-tdTomato;Hes1GFP/+ tumour section showing co-localization of GFP and Tomato signals. f, Flow cytometry shows that GFPhigh cells in pooled tumours from a TKO Rosa26lox-stop-lox-tdTomato;Hes1GFP/+ mouse infected with Ad-CMV-Cre are positive for Tomato expression (representative of n = 2 mice). g, Genotyping PCR analysis for recombination (delta, Δ) or the unrecombined (floxed) alleles at the Rb, p53, and p130 loci in GFPneg and GFPhigh tumour cells sorted from three TKO Hes1GFP/+ mice. DNA from fl/fl and/or Δ/Δ cells served as controls. Scale bars, 50 μm.
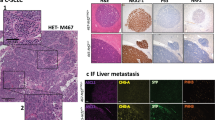
Extended Data Figure 2 Hes1-positive SCLC cells are Notch-pathway-active, epithelial, and do not express neuroendocrine markers.
a, Representative IHC for Hes1 and Notch2 in serial TKO tumour sections. b, RNA-sequencing data of human SCLC tumours (from ref. 8), with positive (ASCL1 and SOX2) and negative (CDX2, MYOD1) controls for expression in SCLC (n = 81 tumours). c–g, TKO Hes1GFP/+ mice were treated with DMSO control (n = 5 mice) or DBZ (n = 4 mice). c, Representative flow cytometry of GFP in pooled tumours from one mouse per condition. Dotted lines delineate the GFPhigh population. d, Quantification of relative median GFP intensity of GFPhigh cells. e, qRT–PCR of sorted GFPneg and GFPhigh cells. f, Immunoblot analysis of sorted GFPneg and GFPhigh cells. At these exposure levels, GFP is detected but not Hes1 in DBZ-treated GFPhigh cells, possibly owing to different half-lives of the proteins (>24 h for GFP53 compared with <1 h for Hes1 (refs 54, 55)). For Notch, an antibody that detected Notch1 cleaved specifically before V1744 (Notch1 ICD), or antibodies that detected the transmembrane + intracellular (excluding the extracellular region, ΔECD) regions of Notch1 or Notch2 were used. g, Representative IHC in treated tumours. h, Flow cytometry of two GFPhigh cell lines (n = 2 biological replicates) grown on dishes coated with Dll4 or 72 h after removal from Dll4. Negative control: GFPneg cell line. i, Quantification of relative median GFP intensity from h (n = 2 biological replicates). j, Immunofluorescence for GFP and NE markers (CGRP and Syp) in TKO Hes1GFP/+ tumours. k, Flow cytometry of the NE marker Ncam1 and GFP in a TKO Hes1GFP/+ tumour (representative of n = 3 biological replicates). The quadrants delineate the negative gates for the GFP and Ncam1 signals and show that GFPhigh cells are mostly negative for Ncam1 expression. l, qRT–PCR analysis of NE genes in GFPhigh cells relative to GFPneg cells (sorted from pooled tumours from n = 3 mice, with n = 3 technical replicates each). m, Flow cytometry analysis of the epithelial marker EpCam and GFP (left) or CD44 and GFP (right) in a TKO Hes1GFP/+ tumour (representative of n = 3 biological replicates). n, qRT–PCR of vimentin (Vim) in GFPneg and GFPhigh cells (n = 4 mice). *P < 0.05; **P < 0.01. Statistical significance was determined by two-tailed unpaired (d, e) or paired (i, l, n) Student’s t-test. Data are represented as mean ± s.d. Scale bars, 50 μm.
Extended Data Figure 3 Notch-active SCLC cells have switched to a non-neuroendocrine fate.
a, Principal component analysis of normalized microarray gene expression values of GFPneg and GFPhigh tumour cells sorted from three TKO Hes1GFP/+ mice. The first two principal components (PCs) accounting for 77.6% of the total variance are shown. b, Heatmap for differentially expressed genes in GFPneg and GFPhigh tumour cells by microarray. c, Gene set enrichment analysis for a Notch pathway signature enriched in GFPhigh cells. d, Gene set enrichment analysis for a neuronal gene set (from the MSigDB C2 curated gene sets collection) enriched in GFPneg cells. e, Gene set enrichment analysis of differentially expressed genes in GFPneg and GFPhigh cells. Gene sets enriched at FDR q value < 0.25 in the MSigDB C5 cellular component ontology are plotted as a function of normalized enrichment score against the q value. Left: gene sets enriched in GFPneg cells; gene sets related to neuronal signatures are highlighted in purple. Right: gene sets enriched in GFPhigh cells; gene sets related to the extracellular region are highlighted in green. f, g, Enrichr analysis for cellular component (f) or biological process (g) Gene Ontology terms significantly enriched in GFPneg or GFPhigh cells. The top ten Gene Ontology terms ranked by combined score are shown. h, Enrichr analysis for cell lines in the Cancer Cell Line Encyclopedia that closely resemble GFPneg or GFPhigh cells. The top ten Gene Ontology terms ranked by combined score are shown. MTC, medullary thyroid carcinoma, an NE tumour of the thyroid; AC, adenocarcinoma.
Extended Data Figure 4 Non-neuroendocrine, Notch-active cells in SCLC tumours are slower-growing than the neuroendocrine cells.
a, b, Co-immunofluorescence for GFP and EdU (a) or phospho-histone H3 (b) in TKO Hes1GFP/+ tumours. c, Weight of tumours formed from freshly sorted GFPneg and GFPhigh tumour cells implanted subcutaneously in immunocompromised NSG mice (n = 6 tumours each; two-tailed unpaired Student’s t-test). d, Flow cytometry of GFP in a tumour formed from freshly sorted GFPneg cells implanted subcutaneously in immunocompromised NSG mice (representative of n = 6 biological replicates). *P < 0.05. Data are represented as mean ± s.d. Scale bars, 50 μm.
Extended Data Figure 5 Non-neuroendocrine, Notch-active cells in SCLC tumours are generated from neuroendocrine tumour cells.
a, Representative IHC for Hes1 and cleaved caspase-3 (CC3) in serial TKO tumour sections. Inset: higher magnification of a positive control for CC3 (tumour from a mouse treated with chemotherapy). b, Predicted numbers and ratios of GFPneg and GFPhigh tumour cells if the two populations divide independently of each other, and GFPneg cells cycle approximately three times faster than GFPhigh cells. c, Representative IHC in serial sections from TKO Hes1GFP/+ tumours initiated by Adeno-CGRP-Cre. d, Quantification of the frequency (Freq.) of Hes1pos cells in TKO hyperplasias (n = 23, from five mice) and tumours (n = 50, from seven mice) induced by Adeno-CGRP-Cre. Scores: 0 (0%), 1 (1–20%), 2 (20–60%), 3 (>60%). e, Flow cytometry analysis showing the percentage of GFPhigh cells in pooled tumours from TKO Hes1GFP/+ mice (n = 4) infected with Ad-CGRP-Cre. f, Images of freshly isolated GFPneg cells grown on dishes coated with Dll4 or PBS control (representative of n = 3 biological replicates). g, h, Freshly isolated GFPneg cells that remained GFPneg after culture on Dll4-coated dishes were re-plated on dishes coated with Dll4 ligand (+Dll4) or PBS control (-Dll4). g, Flow cytometry and images (representative of n = 2 biological replicates). h, GFPneg and GFPhigh cells that formed after this second round of Dll4 stimulation were sorted and analysed by immunoblot. Control: GFPhigh cell line. i, Relative number of GFPhigh cells formed from freshly isolated GFPneg cells grown on Dll4 after 2 weeks of tarextumab treatment (n = 3 biological replicates each). j, Single-cell qRT–PCR (n = 45 each) of H29, H82, and H889 human SCLC cell lines. Heatmap was generated by unsupervised clustering of each cell line. Dark blue regions indicate undetectable expression. k, qRT–PCR for HES1 after 72 h of culture with or without Dll4. Data are normalized to GAPDH (n = 3 biological replicates with n = 3 technical replicates each). l, m, GFPhigh cell lines were treated with DMSO or DBZ while grown with or without Dll4 or co-cultured with three individual NE cell lines in the absence of Dll4. GFP expression was analysed by flow cytometry after 72 h. l, Flow cytometry of GFPhigh cell line 1 (representative of n = 3 biological replicates; GFP intensity quantified in Fig. 2e). m, Quantification of GFP intensity in GFPhigh cell line 2; relative median GFP intensity normalized to the ‘DMSO-Dll4’ condition (n = 3 biological replicates). n, o, Representative images (n) and qRT–PCR (o) of GFPhigh cell lines cultured in the presence of absence of Dll4 for more than a month (n = 3 biological replicates with n = 3 technical replicates). p, Freshly isolated GFPneg cells that became GFPhigh after culture on Dll4 were re-plated on dishes coated with Dll4 ligand or PBS control and analysed by immunoblot after a month. GFPneg cell lysate: positive control for Ascl1. *P < 0.05; **P < 0.01. Statistical significance was determined by two-tailed paired Student’s t-test. Data are represented as mean ± s.d. Scale bars, 50 μm.
Extended Data Figure 6 Analysis of SCLC cells with intermediate levels of activation of the Notch pathway.
a, Flow cytometry showing gates used to sort for tumour cells with negative (neg), intermediate (int), and high levels of GFP from a TKO Hes1GFP/+ mouse (representative of n = 4 biological replicates). b, qRT–PCR for Notch pathway genes and NE genes in sorted cells (n = 4 biological replicates with n = 3 technical replicates each). c–e, Single-cell qRT–PCR of GFPneg, GFPint, and GFPhigh cells sorted from one TKO Hes1GFP/+ tumour (n = 32 cells sorted per group; 20 cells with low expression of housekeeping genes were excluded from the heatmaps). c, Unsupervised clustering segregates the cells into two main groups. d, e, Supervised clustering (d) and analysis (e) of Hes1 mRNA (normalized to Gapdh) shows that the number of GFPint cells with detectable Hes1 levels is intermediate between GFPneg and GFPhigh cells. f, Representative images of freshly isolated cells in culture (without Dll4). Scale bars, 50 μm. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Statistical significance was determined by two-tailed paired Student’s t-test. Data are represented as mean ± s.d.
Extended Data Figure 7 Upregulation of Rest is required for the NE to non-NE transition.
a, b, Images (a) and immunoblot analysis (b) of a mouse NE SCLC cell line (KP1) ~1 month after N1ICD transduction (representative of n = 2 biological replicates). c, Immunoblot of KP1 cells transduced with Ascl1 shRNAs or a shGFP control. Note that the level of Ascl1 downregulation is similar to that achieved by N1ICD overexpression at a similar time point (not shown). d, qRT–PCR of NE genes in KP1 cells transduced with Ascl1 shRNAs (n = 3 biological replicates with n = 3 technical replicates each). e, Representative images of KP1 cells approximately one month after Ascl1 knockdown. f, qRT–PCR for Rest in GFPneg (n = 3) and GFPhigh (n = 4) cell lines (n = 3 technical replicates each). g, qRT–PCR of freshly isolated GFPneg cells 2 weeks after culture in the absence or presence of Dll4 (n = 2 biological replicates with n = 3 technical replicates each). h, Sanger sequencing verification of n = 4 Rest knockout (Rest KO) KP1 clones, with the sequence of both alleles shown. i, qRT–PCR for NE genes in Rest wild-type (n = 3) and Rest knockout (n = 4) clones (n = 3 technical replicates each). j, Schematic for assay to assess the ability of N1ICD to induce adherent non-NE cells from NE SCLC cells. NE SCLC cells (KP1) were transduced with a N1ICD retrovirus or an empty vector control. Seventy-two hours later, 1,200 cells were seeded per well of a 96-well plate. The number of adherent cells per well was counted after 2 weeks. k, KP1 cells stably integrated with Rest shRNAs or an shGFP control were transducted with an N1ICD retrovirus and the number of adherent cells counted after 2 weeks (see schematic in j) (n = 3 biological replicates with n = 3 technical replicates each). l, qRT–PCR for NE genes in KP1 cells 48 h after Rest overexpression (n = 3 biological replicates with n = 3 technical replicates each). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Statistical significance was determined by two-tailed paired (d, k, l) or unpaired (f) Student’s t-test. Data are represented as mean ± s.d. Scale bars, 50 μm.
Extended Data Figure 8 Notch-active SCLC cells promote the survival of neuroendocrine SCLC cells.
a, Gene set enrichment analysis signatures enriched in GFPhigh (astroglial signature) and GFPneg (neuronal signature) tumour cells on the basis of microarray data of tumours from n = 3 mice. b, Images of GFPneg, GFPhigh, or bulk tumour cells (mixture of GFPneg and GFPhigh) 7 days after isolation from TKO Hes1GFP/+ mice and seeded at equal numbers in 50% Matrigel (representative of n = 3 biological replicates). c, Immunofluorescence for Uchl1 (NE marker) and GFP 10 days after the cells were sorted from TKO Hes1GFP/+ tumours and in 50% Matrigel. Note: most of the spheroids in the bulk culture were composed of only one type of cell (Uchl1posGFPneg or Uchl1negGFPpos); the mixed spheroids shown here are the minority. d, Quantification of the number of each type of spheroids in each culture condition from c (n = 3 mice, two sections each for immunostaining). e–g, Relative luciferase activity of a luciferase-labelled mouse SCLC NE cell line seeded alone or co-cultured with GFPhigh cell lines (n = 2 (e) or n = 3 (f, g) biological replicates with n = 3 technical replicates each). e, KP2 NE cell line, 2% serum. f, g, KP1 (f) and KP2 (g) cell lines, 10% serum. h–j, As in e–g, but the cells were fixed and collected for EdU analysis by flow cytometry (n = 2 (h) or n = 3 (i, j) biological replicates). k, Representative images of KP1 cells seeded in conditioned medium (CM) (2% serum) from KP1 cells or from GFPhigh cell lines. l, Relative luciferase activity of luciferase-labelled KP2 cells 72 h after seeding in CM (2% serum) from KP2 cells or from GFPhigh cell lines (n = 3 biological replicates with n = 3 technical replicates each). m, As in l, but the cells were fixed and collected for EdU analysis by flow cytometry (n = 3 biological replicates). n, o, As in l, but with KP1 (n) or KP2 cells (o) in 10% serum (n = 3 biological replicates with n = 3 technical replicates each). p, q, AlamarBlue cell viability assay for NE SCLC cell lines 72 h after culture with the indicated recombinant proteins (n = 3 biological replicates with n = 3 technical replicates each). r, ELISA assay for midkine in supernatant from NE (n = 4) and GFPhigh (n = 4) cell lines (n = 2 biological replicates each). s, Luminex assay for midkine in serum plasma from control (n = 11) and SCLC (n = 15) patients. t, Relative luciferase activity of luciferase-labelled mouse SCLC NE cell lines (n = 4) 72 h after seeding alone or co-cultured with GFPhigh cell lines and treated with 10 μM DBZ or DMSO control. Data are normalized to NE-monoculture with DMSO (n = 3 biological replicates with n = 3 technical replicates each). *P < 0.05; **P < 0.01; ***P < 0.001. Statistical significance was determined by two-tailed paired Student’s t-test (except for r, s: two-tailed unpaired Student’s t-test). Data are represented as mean ± s.d. Scale bars, 50 μm.
Extended Data Figure 9 Notch-active SCLC cells are more resistant to chemotherapy than neuroendocrine SCLC cells.
a, b, Average cell viability (MTT assay) of NE (n = 4) and GFPhigh (n = 3) cell lines 48 h after cisplatin (a) or etoposide (b) treatment (n = 3 biological replicates with n = 3 technical replicates each). c, d, Quantification of cells expressing cleaved caspase-3 (CC3) (c) and Hes1 (d) in tumours in TKO mice acutely treated with cisplatin and etoposide (CC3: n = 49; Hes1: n = 41 tumours) or a vehicle control (CC3: n = 48; Hes1: n = 67 tumours) (data from two groups of three mice previously described in Figure 4a, b in ref. 39 but analysed as number of CC3pos per tumour, not per mouse). e, Representative immunofluorescence of tumours from TKO Hes1GFP/+ mice acutely treated with cisplatin and etoposide. Scale bars, 50 μm. f, Quantification of Hes1pos cells in the tumours of TKO Rosa26lox-stop-lox-Luciferase mice after 3 weeks of weekly saline (n = 190 tumours, from seven mice) or cisplatin (n = 217 tumours, from eight mice) treatments. g–i, Overall survival of all (g; n = 65, 49 HES1neg, 16 HES1pos), stages I–III (h; n = 26, 19 HES1neg, 7 HES1pos), or stage IV (i; n = 39, 30 HES1neg, 9 HES1pos) patients with SCLC with HES1-negative or HES1-positive tumours. j–l, Progression-free survival of all (j; n = 67, 51 HES1neg, 16 HES1pos), stages I–III (k; n = 28, 21 HES1neg, 7 HES1pos), or stage IV (l; n = 39, 30 HES1neg, 9 HES1pos) patients with SCLC with HES1-negative or HES1-positive tumours. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Statistical significance was determined by two-tailed paired (a, b) or unpaired (c, d, f) Student’s t-test. For g–l, probability was calculated using a weighted log-rank test with emphasis on later time points (see Methods). Data are represented as mean ± s.d.
Extended Data Figure 10 Notch2/3 inhibition in vivo reduces the Notch-active cell population in SCLC and improves anti-tumour response in combination with chemotherapy.
a, Schematic for the generation of TKO allografts. A TKO tumour was isolated, digested to form single cells, and implanted subcutaneously in NSG mice. b, Representative haematoxylin and eosin (H&E) staining and IHC for Ascl1 in allograft tumour sections. c, Tumour volumes of TKO allografts treated with the indicated drugs (n = 16 tumours per group, one experiment). The experiment was stopped at day 18 and samples were collected for analysis. d, e, Representative IHC for Hes1 (d) or double IHC for Hes1 (brown) and Ascl1 (pink) (e) in TKO allograft tumours collected 18 days after the start of treatment. f, g, IHC staining quantification for the frequency of Hes1posCC3pos (f) and Hes1posKi67pos (g) cells in mouse TKO allografts after 18 days of treatment (n = 8 tumours per group, except for carboplatin/irinotecan and tarextumab in f (n = 7 tumours each)). h, i, IHC staining quantification for the frequency of HES1posCC3pos (h) and HES1posKI67pos (i) cells in xenograft tumours after 31 days of treatment (for h, control/tarextumab groups: n = 4 tumours each; carboplatin/irinotecan: n = 3 tumours; tarextumab + carboplatin/irinotecan: n = 2 tumours; for i, control/tarextumab groups: n = 4 tumours each; carboplatin/irinotecan and tarextumab + carboplatin/irinotecan groups: n = 5 tumours each). j, k, IHC staining quantification for the frequency of Ascl1posCC3pos (j) and Ascl1posKi67pos (k) cells in mouse TKO allografts after 18 days of treatment (n = 8 tumours per group, except for the carboplatin/irinotecan group in j (n = 7 tumours). l, m, IHC staining quantification for the frequency (%) of ASCL1posCC3pos (l) and ASCL1posKI67pos (m) cells in xenograft tumours after 31 days of treatment (n = 4 tumours per group, except for control and tarextumab groups in l (n = 2 tumours) and the tarextumab + carboplatin/irinotecan group in m (n = 3 tumours)). n, o, Representative IHC (n) and quantification of HES1pos (o) cells in LU66 patient-derived xenograft tumours collected 56 days after the start of treatment (control: n = 3 tumours; tarextumab: n = 4 tumours; carboplatin/irinotecan and tarextumab + carboplatin/irinotecan: n = 5 tumours per group). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Statistical significance was determined by two-tailed unpaired Student’s t-test. Data are represented as mean ± s.e.m. Scale bars, 50 μm.
Supplementary information
Supplementary Information
This file contains Supplementary Methods and Supplementary Figure 1. The Supplementary Methods include clinical and biological characteristics of SCLC patients analyzed for the survival study, as well as primer sequences for qRT-PCR, ChIP-qPCR and genotyping. Supplementary Figure 1 contains immunoblot source data for Figures 1h, 2c, 3h and Extended Data Figures 2f, 5h, 5p, 7b and 7c. (PDF 498 kb)
Supplementary Table
This file contains Supplementary Table 1 (XLSX 709 kb)
Supplementary Table
This file contains Supplementary Table 2 (XLSX 13 kb)
Supplementary Table
This file contains Supplementary Table 3 (XLSX 17 kb)
Supplementary Table
This file contains Supplementary Table 4 (XLSX 17 kb)
Supplementary Table
This file contains Supplementary Table 5 (XLSX 14 kb)
Rights and permissions
About this article
Cite this article
Lim, J., Ibaseta, A., Fischer, M. et al. Intratumoural heterogeneity generated by Notch signalling promotes small-cell lung cancer. Nature 545, 360–364 (2017). https://doi.org/10.1038/nature22323
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature22323
This article is cited by
-
Inhibitor of PD-1/PD-L1: a new approach may be beneficial for the treatment of idiopathic pulmonary fibrosis
Journal of Translational Medicine (2024)
-
Small cells – big issues: biological implications and preclinical advancements in small cell lung cancer
Molecular Cancer (2024)
-
Genetically-engineered mouse models of small cell lung cancer: the next generation
Oncogene (2024)
-
Dynamic phenotypic reprogramming and chemoresistance induced by lung fibroblasts in small cell lung cancer
Scientific Reports (2024)
-
FAK-LINC01089 negative regulatory loop controls chemoresistance and progression of small cell lung cancer
Oncogene (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.