Abstract
Blood and lymphatic vasculatures are intimately involved in tissue oxygenation and fluid homeostasis maintenance. Assembly of these vascular networks involves sprouting, migration and proliferation of endothelial cells. Recent studies have suggested that changes in cellular metabolism are important to these processes1. Although much is known about vascular endothelial growth factor (VEGF)-dependent regulation of vascular development and metabolism2,3, little is understood about the role of fibroblast growth factors (FGFs) in this context4. Here we identify FGF receptor (FGFR) signalling as a critical regulator of vascular development. This is achieved by FGF-dependent control of c-MYC (MYC) expression that, in turn, regulates expression of the glycolytic enzyme hexokinase 2 (HK2). A decrease in HK2 levels in the absence of FGF signalling inputs results in decreased glycolysis, leading to impaired endothelial cell proliferation and migration. Pan-endothelial- and lymphatic-specific Hk2 knockouts phenocopy blood and/or lymphatic vascular defects seen in Fgfr1/Fgfr3 double mutant mice, while HK2 overexpression partly rescues the defects caused by suppression of FGF signalling. Thus, FGF-dependent regulation of endothelial glycolysis is a pivotal process in developmental and adult vascular growth and development.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Eelen, G., de Zeeuw, P., Simons, M. & Carmeliet, P. Endothelial cell metabolism in normal and diseased vasculature. Circ. Res. 116, 1231–1244 (2015)
Simons, M., Gordon, E. & Claesson-Welsh, L. Mechanisms and regulation of endothelial VEGF receptor signalling. Nat. Rev. Mol. Cell Biol. 17, 611–625 (2016)
De Bock, K., Georgiadou, M. & Carmeliet, P. Role of endothelial cell metabolism in vessel sprouting. Cell Metab. 18, 634–647 (2013)
Ornitz, D. M. & Itoh, N. The fibroblast growth factor signaling pathway. Wiley Interdiscip. Rev. Dev. Biol. 4, 215–266 (2015)
Wang, Y. et al. Ephrin-B2 controls VEGF-induced angiogenesis and lymphangiogenesis. Nature 465, 483–486 (2010)
Bazigou, E. et al. Genes regulating lymphangiogenesis control venous valve formation and maintenance in mice. J. Clin. Invest. 121, 2984–2992 (2011)
James, J. M., Nalbandian, A. & Mukouyama, Y. S. TGFβ signaling is required for sprouting lymphangiogenesis during lymphatic network development in the skin. Development 140, 3903–3914 (2013)
Herbert, C. et al. Molecular mechanism of SSR128129E, an extracellularly acting, small-molecule, allosteric inhibitor of FGF receptor signaling. Cancer Cell 23, 489–501 (2013)
De Bock, K. et al. Role of PFKFB3-driven glycolysis in vessel sprouting. Cell 154, 651–663 (2013)
Zheng, W., Aspelund, A. & Alitalo, K. Lymphangiogenic factors, mechanisms, and applications. J. Clin. Invest. 124, 878–887 (2014)
Patra, K. C. et al. Hexokinase 2 is required for tumor initiation and maintenance and its systemic deletion is therapeutic in mouse models of cancer. Cancer Cell 24, 213–228 (2013)
Srinivasan, R. S. et al. Lineage tracing demonstrates the venous origin of the mammalian lymphatic vasculature. Genes Dev. 21, 2422–2432 (2007)
Cao, R. et al. Mouse corneal lymphangiogenesis model. Nat. Protoc. 6, 817–826 (2011)
Kim, J. W., Gao, P., Liu, Y. C., Semenza, G. L. & Dang, C. V. Hypoxia-inducible factor 1 and dysregulated c-Myc cooperatively induce vascular endothelial growth factor and metabolic switches hexokinase 2 and pyruvate dehydrogenase kinase 1. Mol. Cell. Biol. 27, 7381–7393 (2007)
Wilhelm, K. et al. FOXO1 couples metabolic activity and growth state in the vascular endothelium. Nature 529, 216–220 (2016)
Ichise, T., Yoshida, N. & Ichise, H. FGF2-induced Ras–MAPK signalling maintains lymphatic endothelial cell identity by upregulating endothelial-cell-specific gene expression and suppressing TGFβ signalling through Smad2. J. Cell Sci. 127, 845–857 (2014)
Chen, P. Y. et al. FGF regulates TGF-β signaling and endothelial-to-mesenchymal transition via control of let-7 miRNA expression. Cell Reports 2, 1684–1696 (2012)
Oladipupo, S. S. et al. Endothelial cell FGF signaling is required for injury response but not for vascular homeostasis. Proc. Natl Acad. Sci. USA 111, 13379–13384 (2014)
Murakami, M. et al. The FGF system has a key role in regulating vascular integrity. J. Clin. Invest. 118, 3355–3366 (2008)
Rash, B. G., Lim, H. D., Breunig, J. J. & Vaccarino, F. M. FGF signaling expands embryonic cortical surface area by regulating Notch-dependent neurogenesis. J. Neurosci. 31, 15604–15617 (2011)
de Alboran, I. M. et al. Analysis of c-Myc function in normal cells via conditional gene-targeted mutation. Immunity 14, 45–55 (2001)
Muzumdar, M. D., Tasic, B., Miyamichi, K., Li, L. & Luo, L. A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605 (2007)
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013)
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011)
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010)
Young, M. D., Wakefield, M. J., Smyth, G. K. & Oshlack, A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11, R14 (2010)
Chittenden, T. W. et al. Therapeutic implications of GIPC1 silencing in cancer. PLoS ONE 5, e15581 (2010)
Chittenden, T. W. et al. nEASE: a method for gene ontology subclassification of high-throughput gene expression data. Bioinformatics 28, 726–728 (2012)
Alves, T. C. et al. Integrated, step-wise, mass-isotopomeric flux analysis of the tca cycle. Cell Metab. 22, 936–947 (2015)
Kibbey, R. G. et al. Mitochondrial GTP regulates glucose-stimulated insulin secretion. Cell Metab. 5, 253–264 (2007)
Wu, R. et al. Reduction in hexokinase II levels results in decreased cardiac function and altered remodeling after ischemia/reperfusion injury. Circ. Res. 108, 60–69 (2011)
Dubrac, A. et al. Targeting NCK-mediated endothelial cell front-rear polarity inhibits neovascularization. Circulation 133, 409–421 (2016)
Tang, Z. et al. A mouse model of the cornea pocket assay for angiogenesis study. J. Vis. Exp. (54) 3077 (2011)
Acknowledgements
We thank R. Adams for Cdh5-CreERT2, T. Makinen for Prox1-CreERT2(BAC), G. Oliver for Prox1-CreERT2(KI), as well as F. Vaccarino and S. Tomasi for Fgfr1flox/flox;Fgfr3+/− mice. We also thank H. Ardehali for providing HK2 adenovirus, B. G. Coon for generating CRISPR–Cas9 lentivirus, K. van den Dries for real-time imaging and analysis of cell motility, G. Eelen and A. Quaegebeur for metabolic analysis, T. Yarovinsky for instruction and assistance on the flow cytometry, A. Kuo for help with the Seahorse metabolic analyser and F. Zhuo for schematic illustrations. P.Y. is funded by an American Heart Association Postdoctoral Fellowship (15POST25830021). J.Z. was funded by the Brown-Coxe Fellowship from Yale University. F.D.S. was supported as a postdoctoral researcher by the Research Foundation Flanders (FWO). P.C. is funded by IUAP P7/03, long-term structural Methusalem funding by the Flemish Government, FWO G.0598.12, G.0532.10, G.0817.11, G.0834.13, 1.5.202.10.N Krediet aan navorsers, Leducq Transatlantic Network ARTEMIS, AXA Research Fund (1465) and Foundation against Cancer, European Research Council (ERC) Advanced Research Grant (EU-ERC269073, PC). M.P. is supported by the Max Planck Society, a ERC Starting Grant ANGIOMET (311546), the Deutsche Forschungsgemeinschaft (SFB 834), the Excellence Cluster Cardiopulmonary System (EXC 147/1), the LOEWE grant Ub-Net, the DZHK (German Center for Cardiovascular Research), and the European Molecular Biology Organization Young Investigator Programme. M.S. is supported by National Institutes of Health grants HL053793, HL084619 and ARTEMIS Leducq Transatlantic Network.
Author information
Authors and Affiliations
Contributions
P.Y. performed most of the experiments and prepared figures. K.W. analysed Myc mutant mice and performed other Myc-related experiments. A.D. performed retina analysis and sprouting assays. J.K.T. performed sprouting assay and western blotting analysis. T.C.A., J.S.F., Y.X., F.D.S. and J.s.Z. performed the mass spectrometry, fluorescence-activated cell sorting analysis, ChIP, SSR studies and cornea lymphangiogenesis assay respectively. J.Z. performed western blotting analysis. Z.C., L.S., H.S. and T.W.C. contributed to RNA-seq analysis. N.H. provided Hk2flox/flox mice. S.-W.J., R.G.K., K.K.H., P.C., A.E. and M.P. discussed results and provided comments. M.S. supervised the project and wrote the manuscript together with P.Y.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks M. Kahn and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
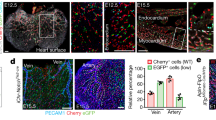
Extended Data Figure 1 Expression of FGFRs in mouse and human LECs and the effect of single knockout of Fgfr1 or Fgfr3 on lymphatic development in the embryonic skin.
a, b, qPCR analysis of FGFR expression in mouse dermal LECs (isolated from E15.5 embryos by FACS) (a; n = 3 embryos) and HDLECs (b; n = 3 technical replicates, representative of two experiments). c, d, qPCR analysis of FGFR expression in HDLECs with FGFR1 (c) or FGFR3 (d) knockdown. FGFR mRNA levels in FGFR1- or FGFR3-deficient cells were presented as values relative to those of control siRNA-treated cells. n = 6 replicates (two independent experiments, technical triplicates per experiment). e, f, Activation of mTmG reporter by Cdh5-CreERT2 (e; scale bar, 100 μm) or Prox1-CreERT2(BAC) (f; scale bar, 250 μm) in dermal LECs of E15.5 mouse embryos. g, Anterior dorsal skin with VEGFR3 and PECAM1 staining from E15.5 Fgfr1iΔLEC(BAC) and Fgfr1flox/flox embryos, which were treated with tamoxifen at E12.5 and E13.5. e, Arrowheads indicate lymphatic vessels marked by EGFP. Scale bar, 250 μm. h, Quantification of the distance between the two leading fronts of the lymphatic vessels (n = 6 embryos for Fgfr1flox/flox; n = 5 embryos for Fgfr1iΔLEC(BAC)). i, Anterior dorsal skin stained for VEGFR3 and PECAM1 from E15.5 Fgfr1flox/flox;Fgfr3+/+, Fgfr1flox/flox;Fgfr3+/− and Fgfr1flox/flox;Fgfr3−/− embryos. Scale bar, 250 μm. j, Quantification of the distance between the two leading fronts of the lymphatic vessels (n = 4 embryos for Fgfr1flox/flox;Fgfr3+/+; n = 6 embryos for Fgfr1flox/flox;Fgfr3+/−; n = 2 embryos for Fgfr1flox/flox;Fgfr3−/−). Note that these embryos were not treated with tamoxifen. Dotted lines indicate the midline in g and i. Data represent mean ± s.e.m., ***P < 0.001, NS is non-significant, calculated by unpaired t-test (c, d, h) and one-way ANOVA plus Tukey’s multiple comparisons test (j).
Extended Data Figure 2 Endothelial Fgfr1/Fgfr3 deletion impairs dermal lymphatic development and inhibition of FGF signalling suppresses pathological lymphangiogenesis.
a, Experimental strategy. b, Bright-field images of E15.5 Fgfr1iΔEC;Fgfr3−/− and control (Fgfr1flox/flox;Fgfr3+/−) embryos with tamoxifen treatment at E10.5 and E11.5. Arrowhead denotes area with lymphoedema. c, Representative images of anterior dorsal skin with VEGFR3 and PECAM1 staining from E15.5 Fgfr1iΔEC;Fgfr3−/− and control embryos treated with tamoxifen at E10.5 and E11.5. d, e, Quantification of the distance between the two leading fronts of the lymphatic vessels (d; n = 3 litters) and the number of lymphatic branch points per square millimetre of skin area (e; n = 9 embryos for control (Fgfr1flox/flox;Fgfr3+/−); n = 7 embryos for Fgfr1iΔEC;Fgfr3−/−). f, Experimental strategy. g, Representative images of anterior dorsal skin with VEGFR3 and PECAM1 staining from E15.5 Fgfr1iΔEC;Fgfr3−/− and control embryos with tamoxifen treatment at E11.5 and E12.5. h, i, Quantification of the distance between the two leading fronts of the lymphatic vessels (h; n = 4 litters) and the number of lymphatic branch points per square millimetre of skin area (i; n = 9 embryos for control (Fgfr1flox/flox;Fgfr3+/−); n = 8 embryos for Fgfr1iΔEC;Fgfr3−/−). In c, g, bottom panels (scale bar, 100 μm) for each genotype are high-magnification images of boxed regions in the top panels (scale bar, 250 μm). Double-headed arrows indicate the distance between the two leading fronts of the lymphatic vessels, which is larger in Fgfr1iΔEC;Fgfr3−/− skin than in control. j, Representative images of the peri-tumoural area of orthotopic Panc02 tumours stained for LYVE1 after vehicle (control) or FGFR-inhibitor treatment (SSR). k, Quantification of the area of LYVE1+ lymphatics per peri-tumoural area (n = 3 mice for each condition). Data represent mean ± s.e.m., *P < 0.05, ***P < 0.001, calculated by unpaired t-test (d, e, h, i, k).
Extended Data Figure 3 Endothelial Fgfr1/Fgfr3 are essential for blood vessel development in the embryonic skin.
a, Schematic of the experimental strategy. b, Representative images of anterior dorsal skin with PECAM1 staining from E15.5 Fgfr1iΔEC;Fgfr3−/− and control embryos. Scale bar, 250 μm. c–e, Quantification of the number of blood vessel branch points per square millimetre of skin area (c; n = 9 embryos for control (Fgfr1flox/flox;Fgfr3+/−); n = 7 embryos for Fgfr1iΔEC;Fgfr3−/−), blood vessel covered area relative to skin area (d; n = 9 embryos for control (Fgfr1flox/flox;Fgfr3+/−); n = 7 embryos for Fgfr1iΔEC;Fgfr3−/−) and capillary diameter (e; n = 6 embryos for control (Fgfr1flox/flox;Fgfr3+/−); n = 3 embryos for Fgfr1iΔEC;Fgfr3−/−). f, Anterior dorsal skin stained for Cx40 in E15.5 Fgfr1iΔEC;Fgfr3−/− and control embryos. Scale bar, 250 μm. g, h, Quantification of the number of artery branch points (g) and artery diameter (h). n = 6 embryos for control (Fgfr1flox/flox;Fgfr3+/−); n = 3 embryos for Fgfr1iΔEC;Fgfr3−/−. Data represent mean ± s.e.m., *P < 0.05, **P < 0.01, ***P < 0.001, calculated by unpaired t-test (c–e, g, h).
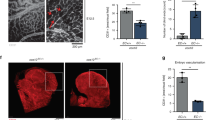
Extended Data Figure 4 Requirement of endothelial Fgfr1/Fgfr3 for retinal angiogenesis.
a, Schematic of the experimental strategy to assess early formation of the retinal vasculature. The red triangles indicate the intragastric injections of tamoxifen at P0, P1 and P2. b, Representative images of isolectin B4 (IB4)-stained retinal vessels in P5 Fgfr1iΔEC;Fgfr3−/− and control mice. Scale bar, 500 μm. c, Quantification of vascular progression (d is the distance between the vascular front and the optic nerve; D is the retina radius), vascular density (AU, arbitrary units) and the number of branch points per square millimetre of retina area. n = 16 retinas for control (Fgfr1flox/flox;Fgfr3−/−); n = 19 retinas for Fgfr1iΔEC;Fgfr3−/−. d, Angiogenic fronts of IB4- and Erg1/2/3-stained retinal vessels in P5 Fgfr1iΔEC;Fgfr3−/− and control mice. Scale bar, 50 μm. e, Quantification of the number of tip cells per 200-μm length (n = 6 retinas for control (Fgfr1flox/flox;Fgfr3−/−); n = 4 retinas for Fgfr1iΔEC;Fgfr3−/−) and the number of vascular front endothelial cells (ECs) per 0.04-mm2 retina area (n = 4 retinas for control (Fgfr1flox/flox;Fgfr3−/−); n = 4 retinas for Fgfr1iΔEC;Fgfr3−/−). f, Retinal vessels stained for phospho-Histone H3 (PH3) and IB4 in P5 Fgfr1iΔEC;Fgfr3−/− and control mice. Scale bar, 50 μm. g, Quantification of the number of PH3+IB4+ endothelial cells per vascular area (normalized to control mice; n = 4 for control (Fgfr1flox/flox;Fgfr3−/−); n = 6 for Fgfr1iΔEC;Fgfr3−/−). h, Representative images of IB4-stained retinal vessels in P5 Fgfr1iΔEC and Fgfr1flox/flox mice. Scale bar, 500 μm. i, Quantification of vascular density and the number of branch points per square millimetre of retina area (n = 18 retinas for Fgfr1flox/flox; n = 8 retinas for Fgfr1iΔEC). j, Representative images of IB4-stained retinal vessels in P5 Fgfr1flox/flox;Fgfr3+/− and Fgfr1flox/flox;Fgfr3−/− mice. Scale bar, 500 μm. k, Quantification of vascular density and the number of branch points per square millimetre of retina area (n = 16 retinas for Fgfr1flox/flox;Fgfr3+/−; n = 16 retinas for Fgfr1flox/flox;Fgfr3−/−). Data represent mean ± s.e.m., **P < 0.01, ***P < 0.001, calculated by unpaired t-test (c, e, g, i, k).
Extended Data Figure 5 Proliferation and migration of HDLECs with FGFR1 and/or FGFR3 knockdown.
a, Proliferation of HDLECs treated with indicated siRNAs was measured by using xCELLigence (see Methods) (n = 4 wells of samples for each condition; representative of two independent experiments). b, Wound-healing assay to assess the migration of HDLECs transfected with siRNAs as indicated. Red dotted lines outline wound area in the last time-point images of HDLECs with different treatments. c, Wound closure area between the first and last time points was measured and normalized to that of control siRNA-treated HDLECs (n = 8 imaging fields for control siRNA, FGFR1 siRNA and FGFR3 siRNA; n = 7 imaging fields for FGFR1/FGFR3 siRNAs; representative of two independent experiments). Data represent mean ± s.e.m., ***P < 0.001, calculated by one-way ANOVA plus Tukey’s multiple comparisons test (a, c).
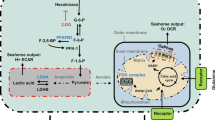
Extended Data Figure 6 Metabolic measurement and glycolytic enzyme expression in HDLECs and HUVECs.
a, Measurement of flux rate of different metabolic processes in HDLECs (glucose oxidation, n = 3 samples; glycolysis, n = 11 samples, combined from three experiments; glutamine oxidation, n = 5 samples, combined from two experiments; and fatty-acid oxidation, n = 4 samples). b, Measurement of glucose uptake in HDLECs treated with or without FGF2 (n = 3 wells of samples for each condition). c, Top 20 protein-coding transcripts (ranked by fold change) that were increased by FGF2 and reduced by FGFR1 siRNA (see Methods for details). HK2, highlighted in red, is the only glucose metabolic gene in this list. d, Western blots showing the knockdown efficiency of HK2 siRNA. e, f, Measurement of glycolytic flux rate of HDLECs with indicated treatments. For e, n = 4 wells of samples for control siRNA, n = 4 wells of samples for HK2 siRNA, n = 3 wells of samples for control siRNA + FGF2, and n = 4 wells of samples for HK2 siRNA + FGF2. For f, n = 4 wells of samples for each condition. g, Measurement of glycolytic flux rate of HUVECs in the absence or presence of FGF2 (n = 4 wells of samples for each condition, representative of two independent experiments). h, i, Representative western blot analysis (h) and densitometric quantification (i; n = 5 replicates from three experiments) of glycolytic enzyme expression in control or FGF2-treated HUVECs. j, Measurement of glycolytic flux rate of HDLECs treated with or without different growth factors (n = 4 wells of samples for each condition). k, Densitometric quantification (n = 3 independent experiments) of glycolytic enzyme expression in HDLECs in the presence or absence of different growth factors. Data represent mean ± s.e.m., *P < 0.05, **P < 0.01, ***P < 0.001, calculated by unpaired t-test (b, f, g, i) and one-way ANOVA plus Sidak’s (e, j) or Dunnett’s (k) multiple comparisons test. For gel source data, see Supplementary Fig. 1.
Extended Data Figure 7 Role of HK2 in FGF-dependent cellular behaviours.
a, b, Proliferation (a; n = 4 wells of samples for each condition) and migration (b; n = 4 imaging fields for each condition) of HDLECs with indicated siRNAs, which were serum-starved and treated with or without FGF2. c, d, Proliferation (c; n = 4 wells of samples for control siRNA + Ad-control, control siRNA + Ad-HK2 and FGFR1 siRNA + Ad-control; n = 3 wells of samples for FGFR1 siRNA + Ad-HK2) and migration (d; n = 8 imaging fields for each condition) of HDLECs with indicated siRNAs, which were cultured in fully supplemented medium and treated with control or HK2 adenovirus. Proliferation was measured using xCELLigence and migration was analysed through a wound-healing assay (see Methods). Wound closure area between the first time point and the last time point was measured and normalized to that of control siRNA-treated HDLECs (b) or HDLECs treated with control siRNA and control adenovirus (d). Red dotted lines outline wound area in the last time-point images of HDLECs with different treatments. e, f, Representative images and quantification of microcarrier beads coated with HDLECs under treatments as indicated. Total length of LEC sprouts per bead was quantified. e, f, Arrowheads indicate sprouts extending from the beads. For e, n = 14 beads for control siRNA, n = 14 beads for HK2 siRNA, n = 19 beads for control siRNA + FGF2 and n = 25 beads for HK2 siRNA + FGF2. For f, n = 25 beads for each condition. Data represent mean ± s.e.m., **P < 0.01, ***P < 0.001, calculated by one-way ANOVA plus Sidak’s multiple comparisons test (a–f).
Extended Data Figure 8 Effect of endothelium-specific deletion of Hk2 on the lymphatic and blood vessel development in the skin.
a, Schematic of the experimental strategy. b, Bright-field images of E15.5 Hk2iΔEC and control embryos treated with tamoxifen at E12.5 and E13.5. Arrowhead denotes area with lymphoedema. c, Representative images of anterior dorsal skin with VEGFR3 and PECAM1 staining from E15.5 Hk2iΔEC and control (Hk2flox/flox) embryos with tamoxifen treatment at E12.5 and E13.5. Double-headed arrows indicate the distance between the two leading fronts of the lymphatic vessels, which is larger in Hk2iΔEC skin than in control. Lower panels (scale bar, 100 μm) for each genotype are high-magnification images of boxed regions in top panels (scale bar, 250 μm). d, e, Quantification of the distance between the two leading fronts of the lymphatic vessels (d; n = 4 litters) and the number of lymphatic branch points per square millimetre of skin area (e; n = 5 embryos for control (Hk2flox/flox); n = 6 embryos for Hk2iΔEC). f, g, Quantification of the number of blood vessel branch points per square millimetre of skin area (f) and area covered by blood vessels (g). n = 5 embryos for control (Hk2flox/flox); n = 6 embryos for Hk2iΔEC. h, LECs were isolated from E15.5 control (Hk2flox/flox) and Hk2iΔEC embryos and analysed for cell cycle distribution. Percentage of cells in different cell cycle phases was quantified (n = 12 embryos for control (Hk2flox/flox) and n = 9 embryos for Hk2iΔEC). i, The experimental strategy. j, Representative images of anterior dorsal skin with PECAM1 staining from E15.5 Hk2iΔEC and control embryos with tamoxifen treatment at E10.5 and E11.5. Scale bar, 250 μm. k, l, Quantification of the number of blood vessel branch points per square millimetre of skin area (k) and blood vessel covered area relative to skin area (l). n = 4 embryos for control (Hk2flox/flox); n = 7 embryos for Hk2iΔEC. m, Anterior dorsal skin stained for Cx40 in E15.5 Hk2iΔEC and control embryos treated with tamoxifen at E10.5 and E11.5. Scale bar, 250 μm. n, Quantification of the number of artery branch points (n = 4 embryos for control (Hk2flox/flox); n = 7 embryos for Hk2iΔEC). Data represent mean ± s.e.m., *P < 0.05, **P < 0.01, calculated by unpaired t-test (d, e, f–h, k, l, n).
Extended Data Figure 9 Endothelial Hk2 is required for retinal angiogenesis.
a, Schematic of the experimental strategy to assess early formation of the retinal vasculature (P0–P5). The red triangles indicate the intragastric injections of tamoxifen at P0, P1 and P2. b, Representative images of IB4-stained retinal vessels in P5 Hk2iΔEC and control mice. Scale bar, 500 μm. c, Quantification of vascular progression (d is the distance between the vascular front and the optic nerve; D is the retina radius), vascular density and the number of branch points per square millimetre of retina area. n = 18 retinas for control (Hk2flox/flox) and n = 24 retinas for Hk2iΔEC. d, Angiogenic fronts of IB4- and Erg1/2/3-stained retinal vessels in P5 Hk2iΔEC and control mice. Scale bar, 50 μm. e, Quantification of the number of tip cells per 200-μm length (n = 4 retinas for control (Hk2flox/flox); n = 4 retinas for Hk2iΔEC) and the number of vascular front ECs per 0.04-mm2 retina area (n = 4 for control (Hk2flox/flox); n = 4 for Hk2iΔEC). f, Retinal vessels stained for PH3 and IB4 in P5 Hk2iΔEC and control mice. Scale bar, 50 μm. g, Quantification of the number of PH3+IB4+ ECs per vascular area (normalized to control mice; n = 4 for control (Hk2flox/flox); n = 4 for Hk2iΔEC). h, Staining for collagen IV (Col4) and IB4 in the retinas of P5 Hk2iΔEC and control mice. Scale bar, 50 μm. i, Quantification of Col4+ area per IB4+ area (n = 6 retinas for control (Hk2flox/flox); n = 8 retinas for Hk2iΔEC). Data represent mean ± s.e.m., **P < 0.01, ***P < 0.001, calculated by unpaired t-test (c, e, g, i).
Extended Data Figure 10 Characterization of FGF–MYC–HK2 signalling in endothelial cells.
a, Schematic showing that E-boxes, which are MYC binding elements, localize in the first intron of human and mouse HK2 genes. Primers were designed to amplify the E-box-containing region (green bar) in ChIP–qPCR assay. b, ChIP–qPCR analysis of DNA immunoprecipitated with MYC antibody or IgG (n = 3 independent experiments) in HDLECs. c, qPCR analysis of HK2 expression in HDLECs transfected with control siRNA or MYC siRNA (n = 4 experiments). d, qPCR analysis of HK2 mRNA in HDLECs infected with control or MYC adenovirus (n = 6 experiments). e, Glycolytic flux measurement of HDLECs transfected with control siRNA or MYC siRNA (n = 4 wells of samples for each treatment, representative of two independent experiments). f, Extracellular acidification rate (ECAR) in HDLECs transfected with control siRNA or MYC siRNA. n = 6 replicates (three independent experiments, duplicates per experiment). g, Extracellular acidification rate in HDLECs infected with control or MYC adenovirus. n = 4 replicates (two independent experiments, duplicates per experiment). h, Anterior dorsal skin was dissected from E15.5 mouse embryos and immunostained with anti-VEGFR3 and anti-MYC antibodies. Arrowheads indicate LECs with Myc expression. i, Confocal images of anterior dorsal skin with VEGFR3, PROX1 and PECAM1 staining from E15.5 MyciΔLEC(BAC) and control embryos. Scale bar, 250 μm. j, k, Quantification of the distance between the leading fronts of ingrowing lymphatics (j; n = 7 embryos for control (Mycflox/flox); n = 9 embryos for MyciΔLEC(BAC)) and the number of LECs (PROX1 staining) per 100-μm length of lymphatic vessels (k; n = 6 embryos for control (Mycflox/flox); n = 8 embryos for MyciΔLEC(BAC)). l, qPCR analysis of HK2 expression in HUVECs transfected with control siRNA or MYC siRNA (n = 4 experiments). m, n, Representative immunoblot analysis (m) and densitometric quantification (n) of HK2 expression in HUVECs transfected with control siRNA or MYC siRNA (n = 4 independent experiments). o, qPCR analysis of HK2 mRNA in HUVECs infected with control or MYC adenovirus (n = 2–4 experiments). p, qPCR analysis of Hk1 and Hk2 expression in dermal BECs isolated from E15.5 MyciΔEC and control embryos with tamoxifen treatment at E11.5 and E12.5 (n = 2–4 embryos). q, r, Representative western blot (q) and densitometric quantification (r) of MYC expression in HUVECs treated with or without FGF2 (n = 5 replicates from three experiments). s, Representative images showing Myc expression was reduced in retinal vasculature of P5 Fgfr1iΔEC;Fgfr3−/− compared with control (tamoxifen treatment from P0 to P2). Arrowheads indicate retinal BECs with Myc expression. Scale bar, 50 μm. t, Quantification of the percentage of MYC+ retinal endothelial cells in the vascular fronts (n = 4 retinas for each genotype). Data represent mean ± s.e.m., *P < 0.05, **P < 0.01, ***P < 0.001, calculated by unpaired t-test (b–g, j–l, n–p, r, t). For gel source data, see Supplementary Fig. 1.
Supplementary information
Supplementary Figure
This file contains Supplementary Figure 1, the uncropped blots. (PDF 1080 kb)
Source data
Rights and permissions
About this article
Cite this article
Yu, P., Wilhelm, K., Dubrac, A. et al. FGF-dependent metabolic control of vascular development. Nature 545, 224–228 (2017). https://doi.org/10.1038/nature22322
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature22322
This article is cited by
-
Gli1-mediated tumor cell-derived bFGF promotes tumor angiogenesis and pericyte coverage in non-small cell lung cancer
Journal of Experimental & Clinical Cancer Research (2024)
-
Lymphatic vessel: origin, heterogeneity, biological functions, and therapeutic targets
Signal Transduction and Targeted Therapy (2024)
-
Lymphatic vessels in the age of cancer immunotherapy
Nature Reviews Cancer (2024)
-
Research progress of exosomes in the angiogenesis of digestive system tumour
Discover Oncology (2024)
-
Efficacy of recombinant bovine basic fibroblast growth factor to reduce hemorrhage after cervical loop electrosurgical excision procedure
BMC Women's Health (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.